Process Based Models
Bayesian Methods for Ecological and Environmental Modelling
UKCEH Edinburgh
What we will cover
11:30 am-1 pm: Session 6b/7a Process Based Models
- What is a process based model (PBM)?
- Why considering uncertainties in PBM predictions may be important
- Sources of uncertainty in the Bayesian inference of PBMs
- Worked example: Bayesian inference of a simple PBM (VSEM) using BayesianTools
1-2 pm: Lunch break
2-3:30 pm: Session 6b/7a Process Based Models
- Practical 7a: Process Based Models
- Why it is important to represent all sources of uncertainty in Bayesian inference?
- Recap and Summary
What is a process based model? (PBM)
Model structure based on mechanistic process understanding
Often deterministic
Good for extrapolation (eg forecasting) and “what if” type questions
Examples:
- General Circulation Models GCMs (Climate modelling)
- Numerical Weather Prediction Models (Weather forcasting)
- Even Newton’s Laws of Motion
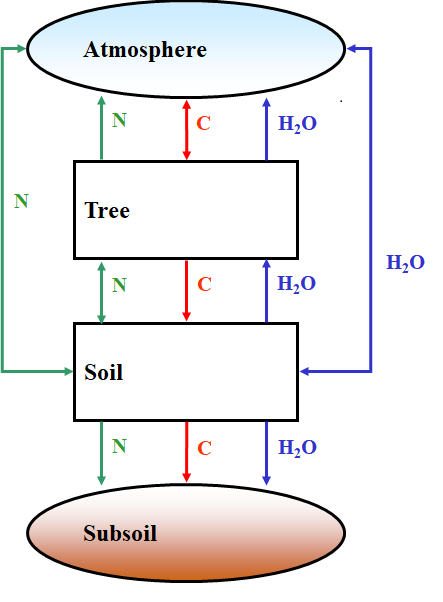
PBM inputs and outputs
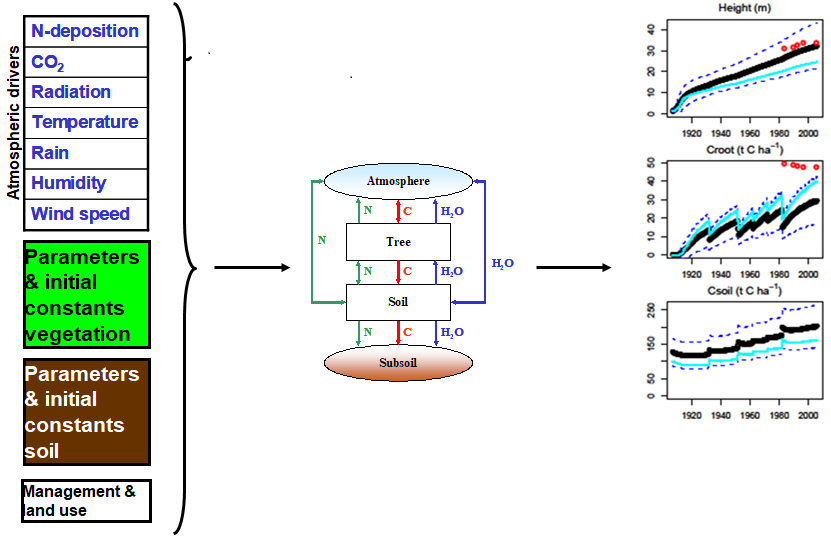
Model Tuning Approach
Aim to understand/predict something in nature
Build a model representing our current best understanding of the processes involved
- Model includes uncertain “tunable” parameters
Collect evidence/data to test/validate model against
Model does not agree with data/evidence
- Try to improve model performance against data by “tuning” parameters
Problems with Model Tuning
Errors in models and data are acknowledged but often ignored
- Data collected is considered ‘truth’ but we know this is wrong
- Tuning an imperfect model against data where errors are ignored often leads to overfitting
Predictions are made without quantifying their reliability
- Not useful for decision making
Does it matter that we ignore uncertainty?
Storm of 1987
“…earlier on today apparently a woman rang the BBC and said she’d heard there was a hurricane on the way. Well if you are watching don’t worry, there isn’t…”
Storm of 1987
“…earlier on today apparently a woman rang the BBC and said she’d heard there was a hurricane on the way. Well if you are watching don’t worry, there isn’t…”
18 died in Britain, four in France
Devastation costs more than one billion pounds
An estimated 15 million trees were lost
Worst storm since 1703, one in 200 year storm for southern Britain
A public enquiry was held and an internal enquiry was conducted by the Met Office

“…unlike 1987, we now run multiple forecasts (called ensembles) that help us to understand the probabilities involved in a forecast and give a better estimate of the risk of high impact weather events.”
Climate Uncertainty
Model predictions without uncertainty could miss less likely but high impact events
Updated process of analysis
Uncertainty in PBMs
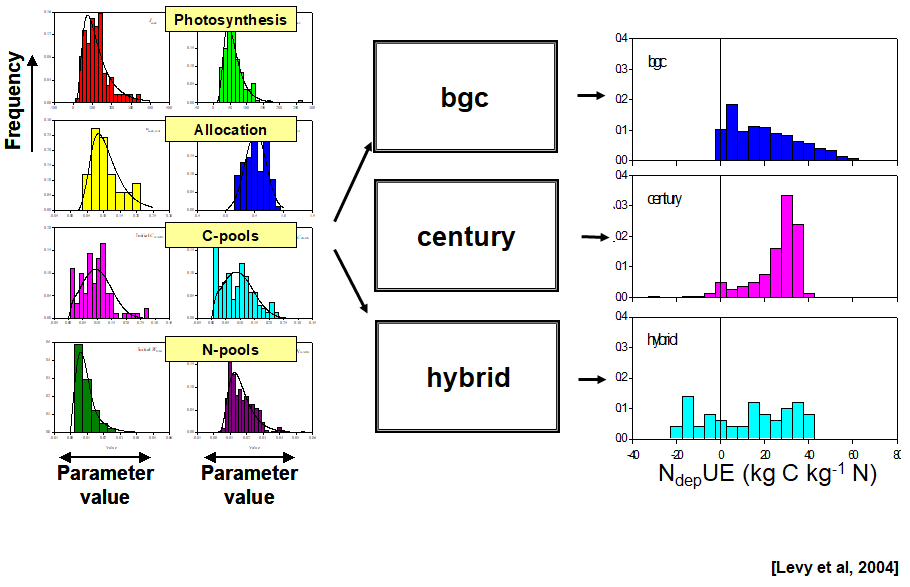
Sources of uncertainty
“random” uncertainty in observations
Variability at scales that we are currently not interested in modelling/predicting.
human or instrument error in taking measurements
representative error
Do observational samples adequately represent the site/system we are interested in - experimental design
Is measurement site (a)typical of (eg pine forest) sites in Europe?
Sources of uncertainty
uncertainty in model driving/input data
Climate/weather data, N deposition, management etc.
model initial value uncertainty
For example which initial soil/tree values are consistent with each other and present day observations
missing or erroneous model processes/structure
All PBMs are wrong
lack of knowledge of model parameter values
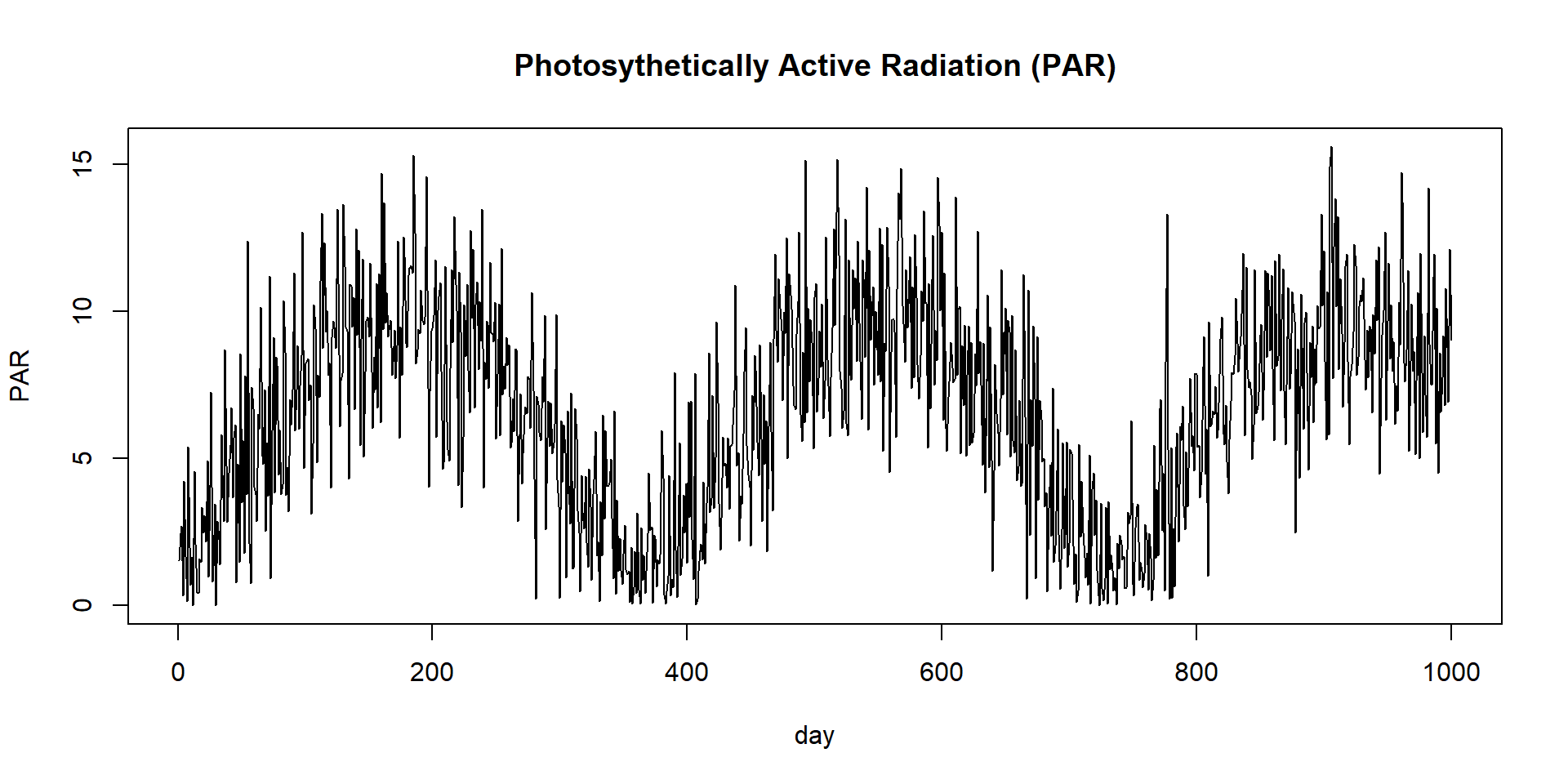
The Very Simple Ecosystem Model (VSEM)
Toy model (designed as part of a theory paper)
Included in BayesianTools (R package that we will use)
Very simplified form of more complex models
To give a concrete example of inferring the parameters of a PBM
VSEM Photosynthesis
Gross/Net primary productivity
\[\begin{align} GPP &= PAR \times LUE \times (1 - \exp^{(-KEXT \times LAI)})\\ NPP &= (1-GAMMA) \times GPP \end{align}\]Net Ecosystem Exchange
\[\begin{align} NEE &= (\frac{C_s}{\tau_s} + GAMMA \times GPP) - GPP \end{align}\]PAR - Photosynthetically active radiation
KEXT - Beer’s law light extinction coeff
LUE - Light use efficiency
LAI - Leaf area index (LAR * Cv)
LAR - Leaf area ratio
Cv/Cs - Vegetative/Soil carbon
GAMMA - Fraction of GPP that is Autotrophic Respiration
VSEM state equations
\[\begin{align} \frac{dC_v}{dt} &= A_v \times NPP &- \frac{C_v}{\tau_v} \\ \frac{dC_r}{dt} &= (1.0-A_v) \times NPP &- \frac{C_r}{\tau_r}\\ \frac{dC_s}{dt} &= \frac{C_r}{\tau_r} + \frac{C_v}{\tau_v} &- \frac{C_s}{\tau_s} \end{align}\]\(C_v\), \(C_r\) and \(C_s\) : Carbon in vegetation, root and soil pools
Run VSEM
| best | lower | upper | |
|---|---|---|---|
| KEXT | 0.500 | 2e-01 | 1e+00 |
| LAR | 1.500 | 2e-01 | 3e+00 |
| LUE | 0.002 | 5e-04 | 4e-03 |
| GAMMA | 0.400 | 2e-01 | 6e-01 |
| tauV | 1440.000 | 5e+02 | 3e+03 |
| tauS | 27370.000 | 4e+03 | 5e+04 |
| tauR | 1440.000 | 5e+02 | 3e+03 |
| Av | 0.500 | 2e-01 | 1e+00 |
| Cv | 3.000 | 0e+00 | 4e+02 |
| Cs | 15.000 | 0e+00 | 1e+03 |
| Cr | 3.000 | 0e+00 | 2e+02 |
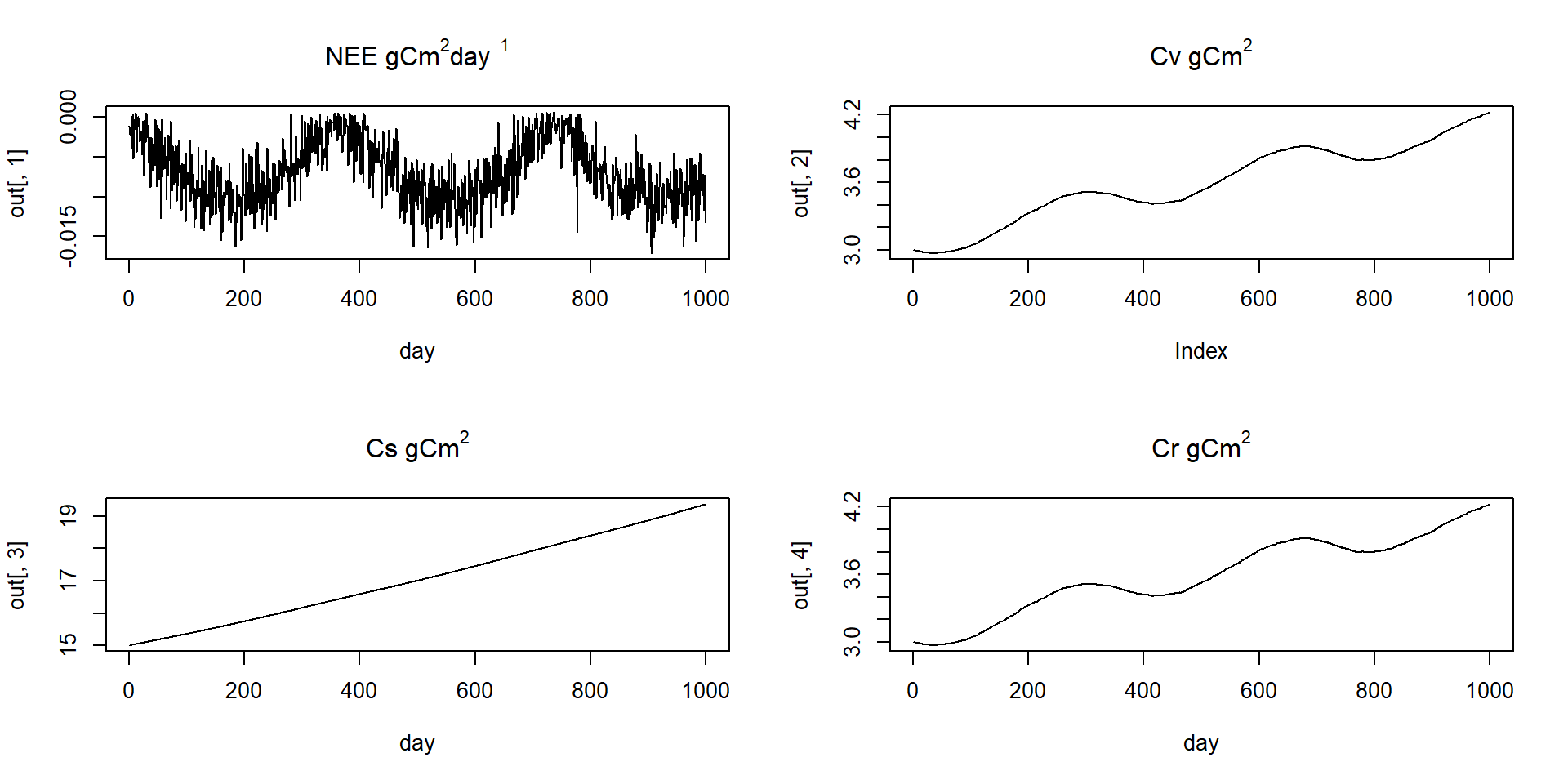
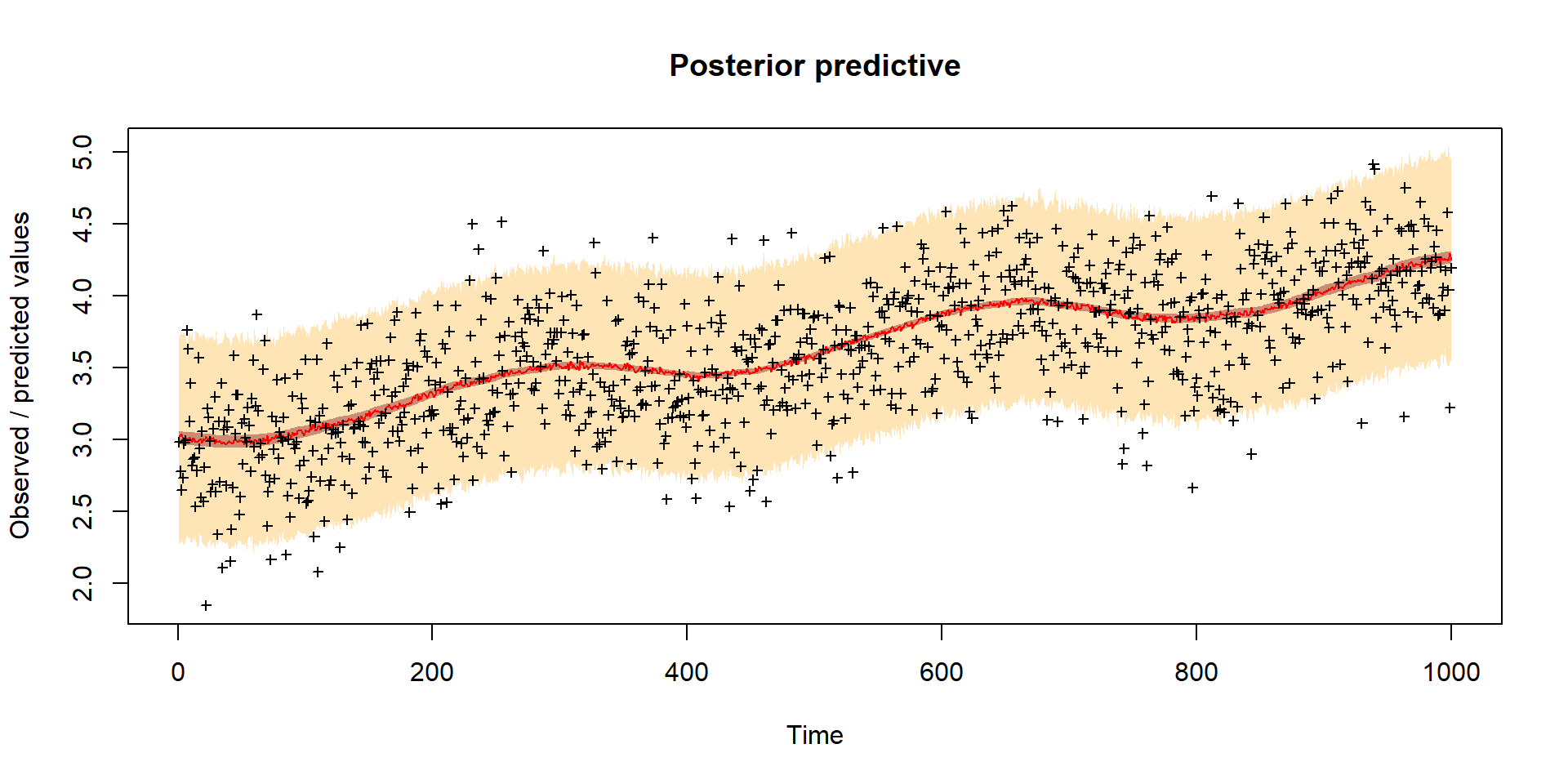
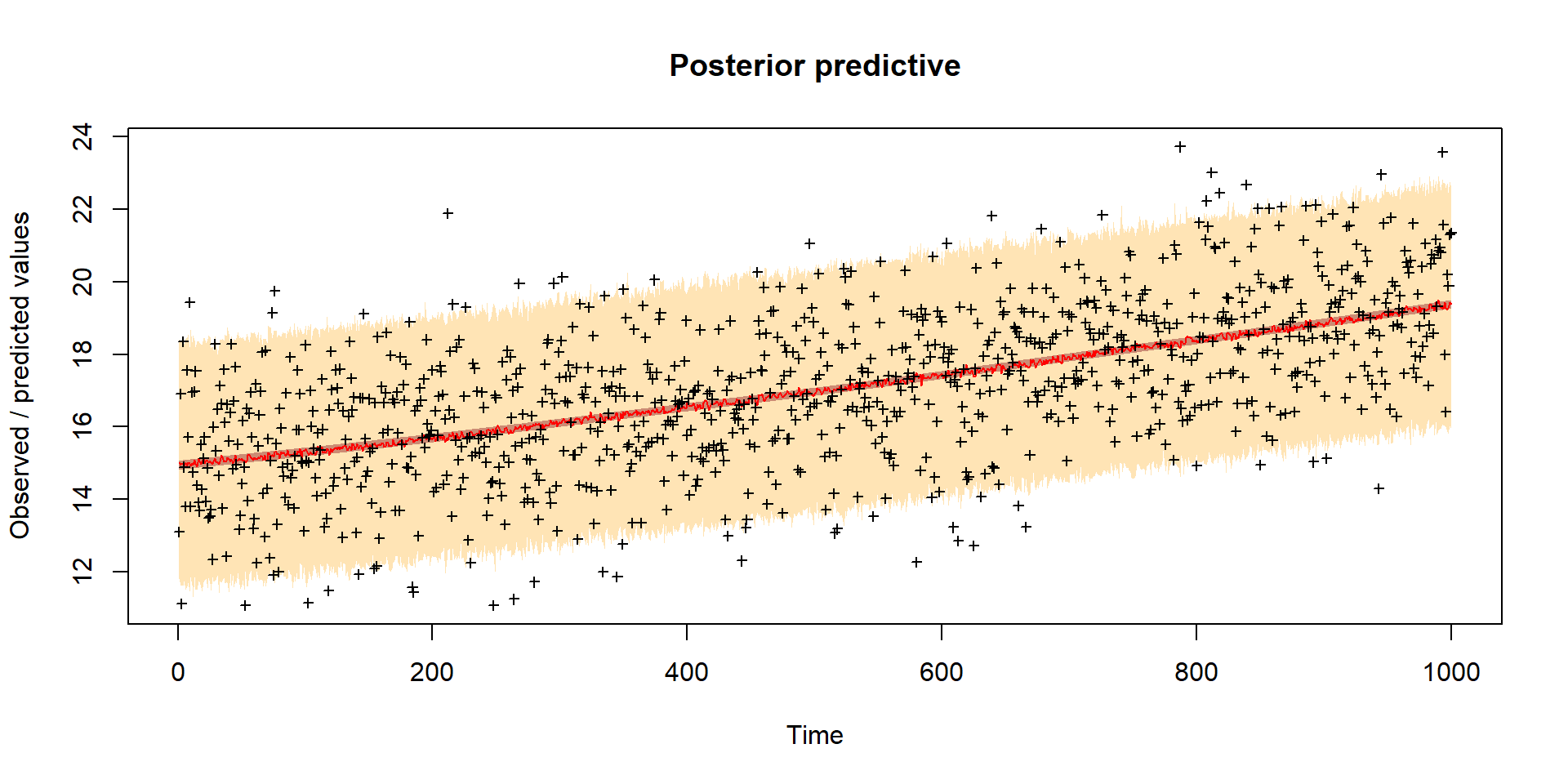
Worked example Bayesian inference of VSEM
Inference of model parameters and initial conditions of carbon pools (Cv, Cs, Cr)
The model structure is fixed i.e. is not being inferred
model inputs and parameters
Create virtual observations
We will not use measured data from the field, but virtual data.
referenceData <- VSEM(refPars$best[1:11], PAR)
referenceData[,1] = 1000 * referenceData[,1]
aobs = referenceData
aobs[,1] = abs(referenceData[,1])
maobs = colMeans(aobs)
obs = referenceData
obs[,1] <- referenceData[,1] + rnorm(length(referenceData[,1]), sd = maobs[1]*refPars$best[12])
obs[,2] <- referenceData[,2] + rnorm(length(referenceData[,1]), sd = maobs[2]*refPars$best[12])
obs[,3] <- referenceData[,3] + rnorm(length(referenceData[,1]), sd = maobs[3]*refPars$best[12])
obs[,4] <- referenceData[,4] + rnorm(length(referenceData[,1]), sd = maobs[4]*refPars$best[12])Why BayesianTools
PBMs often preexist in a language (R, python, FORTRAN, C) Could be costly to recode in an Bayesian package’s bespoke language
- complex models could be hundreds/thousands of lines of code
For PBMs very useful to be able to code a bespoke likelihood function as we need control over calling the PBM
Few MCMC packages allow this (eg BayesianTools, NIMBLE)
Likelihood
Gaussian likelihood Logarithm helps avoid numerical rounding issues
likelihood <- function(par, sum = TRUE){
x = refPars$best
x[parSel] = par
predicted <- VSEM(x[1:11], PAR) # replace here VSEM with your model
predicted[,1] = 1000 * predicted[,1] # this is just rescaling
diff <- c(predicted[,1] - obs[,1])
llValues1 <- dnorm(diff, sd = x[12]*maobs[1], log = T)
diff <- c(predicted[,2] - obs[,2])
llValues2 <- dnorm(diff, sd = x[12]*maobs[2], log = T)
diff <- c(predicted[,3] - obs[,3])
llValues3 <- dnorm(diff, sd = x[12]*maobs[3], log = T)
diff <- c(predicted[,4] - obs[,4])
llValues4 <- dnorm(diff, sd = x[12]*maobs[4], log = T)
return(sum(llValues1,llValues2,llValues3,llValues4))
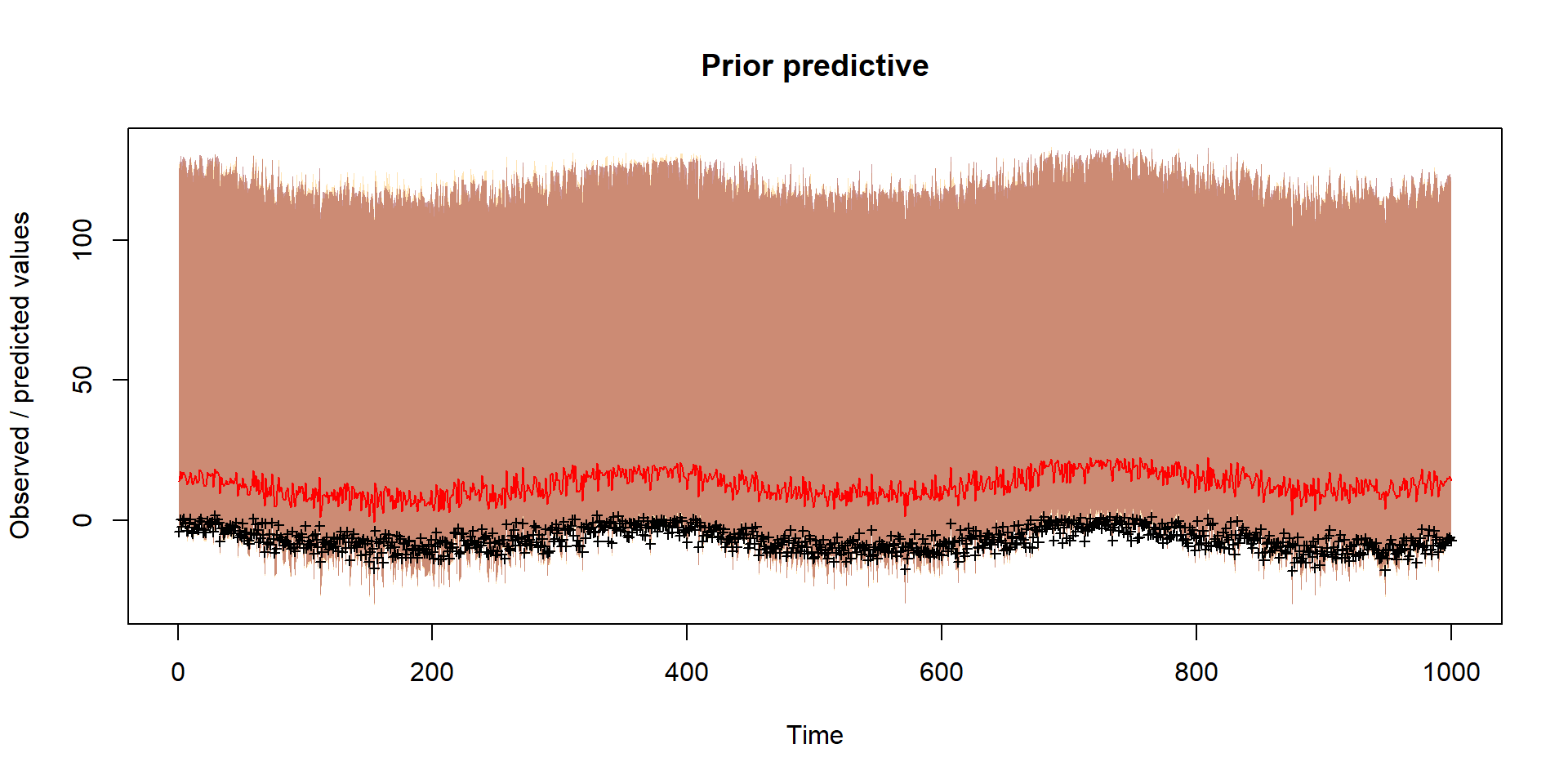
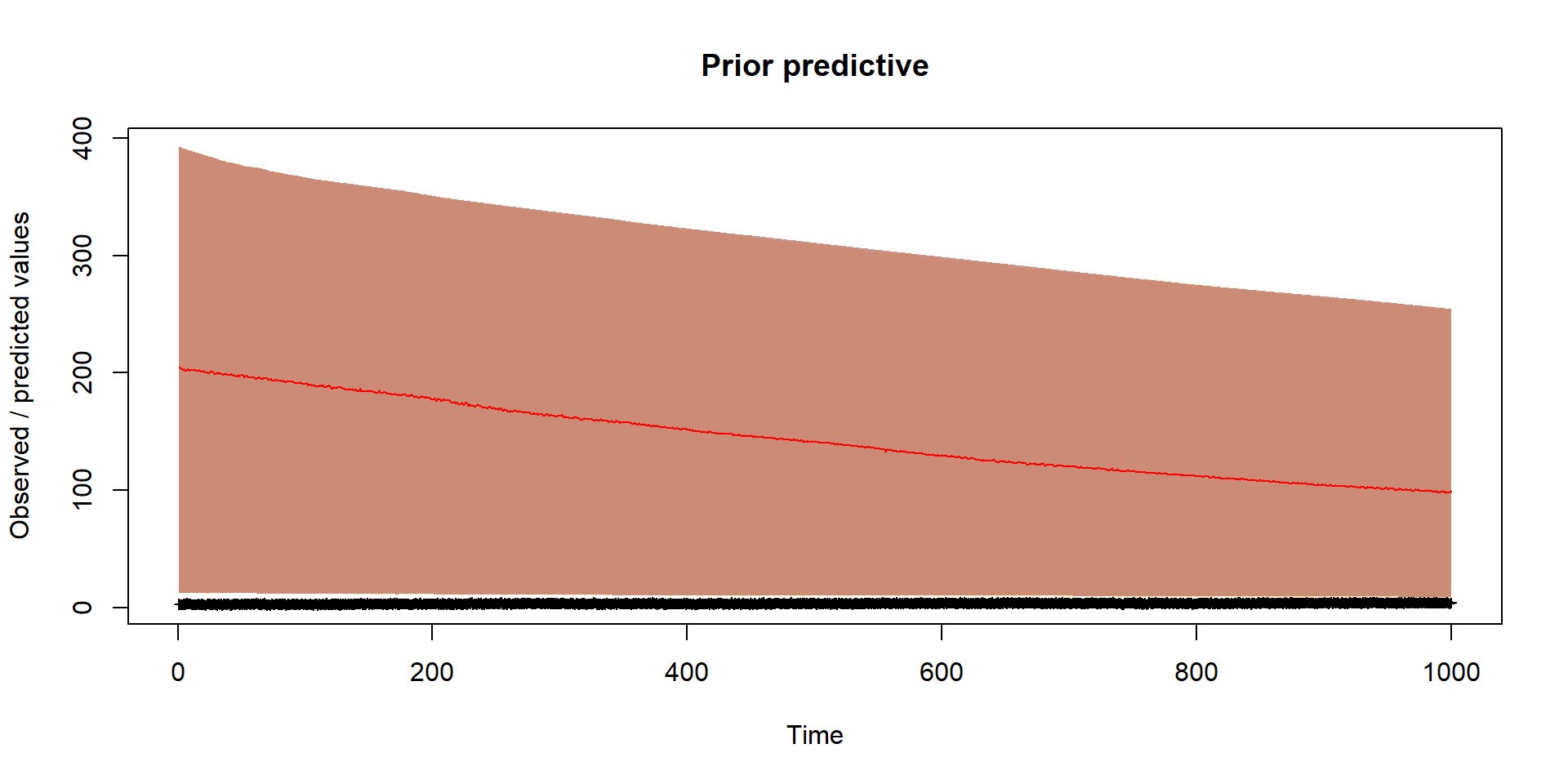
}Prior
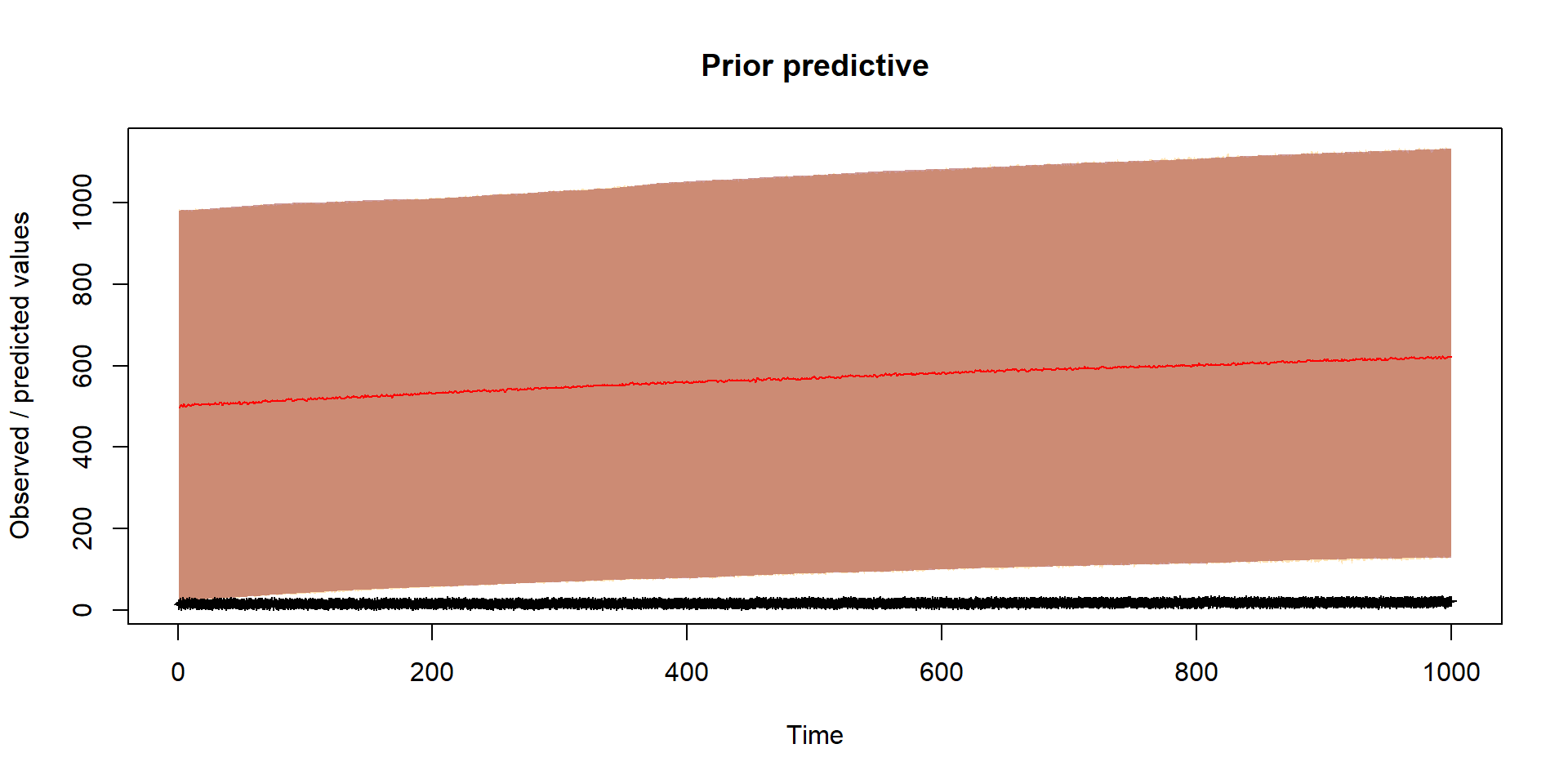
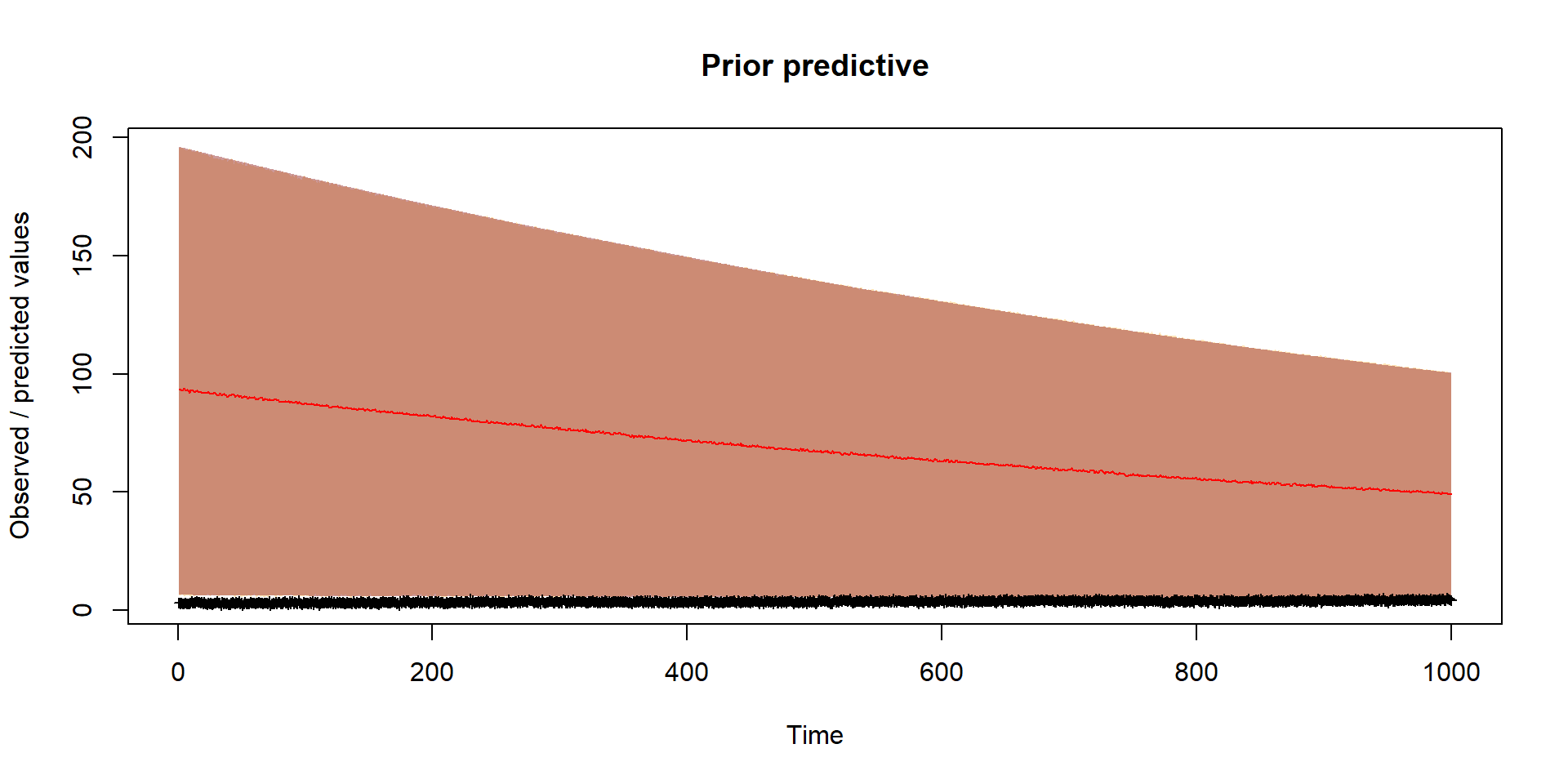
Should reflect prior thinking about uncertainty in parameters
Run inference
More than one chain helps test for convergence
bayesianSetup <- createBayesianSetup(likelihood, prior,
names = rownames(refPars)[parSel])
settings <- list(iterations = 10000, nrChains = 2)
out <- runMCMC(bayesianSetup = bayesianSetup,
sampler = "DEzs", settings = settings)
Running DEzs-MCMC, chain 1 iteration 300 of 10002 . Current logp -548033.4 -652443.7 -167916 . Please wait!
Running DEzs-MCMC, chain 1 iteration 600 of 10002 . Current logp -256349.6 -379429.6 -122381.7 . Please wait!
Running DEzs-MCMC, chain 1 iteration 900 of 10002 . Current logp -54549.21 -66765.04 -114665 . Please wait!
Running DEzs-MCMC, chain 1 iteration 1200 of 10002 . Current logp -53635.39 -66148.04 -49971.64 . Please wait!
Running DEzs-MCMC, chain 1 iteration 1500 of 10002 . Current logp -51801.92 -53685.85 -37625.21 . Please wait!
Running DEzs-MCMC, chain 1 iteration 1800 of 10002 . Current logp -46611.68 -49890.01 -22732.01 . Please wait!
Running DEzs-MCMC, chain 1 iteration 2100 of 10002 . Current logp -34211.66 -9117.365 -7743.43 . Please wait!
Running DEzs-MCMC, chain 1 iteration 2400 of 10002 . Current logp -7509.263 -9117.365 -7502.083 . Please wait!
Running DEzs-MCMC, chain 1 iteration 2700 of 10002 . Current logp -7509.263 -6717.251 -7502.083 . Please wait!
Running DEzs-MCMC, chain 1 iteration 3000 of 10002 . Current logp -5761.173 -5926.76 -6650.44 . Please wait!
Running DEzs-MCMC, chain 1 iteration 3300 of 10002 . Current logp -5761.173 -5926.76 -6395.099 . Please wait!
Running DEzs-MCMC, chain 1 iteration 3600 of 10002 . Current logp -5640.148 -5926.76 -5952.142 . Please wait!
Running DEzs-MCMC, chain 1 iteration 3900 of 10002 . Current logp -5640.148 -5719.07 -5510.251 . Please wait!
Running DEzs-MCMC, chain 1 iteration 4200 of 10002 . Current logp -5640.148 -5719.07 -5510.251 . Please wait!
Running DEzs-MCMC, chain 1 iteration 4500 of 10002 . Current logp -5564.135 -5622.641 -5392.286 . Please wait!
Running DEzs-MCMC, chain 1 iteration 4800 of 10002 . Current logp -5419.243 -5476.586 -5349.391 . Please wait!
Running DEzs-MCMC, chain 1 iteration 5100 of 10002 . Current logp -5250.217 -5446.594 -5349.391 . Please wait!
Running DEzs-MCMC, chain 1 iteration 5400 of 10002 . Current logp -4994.842 -5241.814 -5349.391 . Please wait!
Running DEzs-MCMC, chain 1 iteration 5700 of 10002 . Current logp -4614.035 -5119.363 -5121.21 . Please wait!
Running DEzs-MCMC, chain 1 iteration 6000 of 10002 . Current logp -4474.206 -5115.72 -4961.434 . Please wait!
Running DEzs-MCMC, chain 1 iteration 6300 of 10002 . Current logp -4446.166 -4683.113 -4483.122 . Please wait!
Running DEzs-MCMC, chain 1 iteration 6600 of 10002 . Current logp -4427.97 -4572.002 -4424.495 . Please wait!
Running DEzs-MCMC, chain 1 iteration 6900 of 10002 . Current logp -4423.306 -4439.836 -4366.523 . Please wait!
Running DEzs-MCMC, chain 1 iteration 7200 of 10002 . Current logp -4423.306 -4428.136 -4365.814 . Please wait!
Running DEzs-MCMC, chain 1 iteration 7500 of 10002 . Current logp -4423.306 -4418.397 -4365.814 . Please wait!
Running DEzs-MCMC, chain 1 iteration 7800 of 10002 . Current logp -4398.508 -4418.397 -4365.814 . Please wait!
Running DEzs-MCMC, chain 1 iteration 8100 of 10002 . Current logp -4324.663 -4338.021 -4348.939 . Please wait!
Running DEzs-MCMC, chain 1 iteration 8400 of 10002 . Current logp -4282.213 -4316.847 -4345.466 . Please wait!
Running DEzs-MCMC, chain 1 iteration 8700 of 10002 . Current logp -4262.632 -4316.847 -4345.946 . Please wait!
Running DEzs-MCMC, chain 1 iteration 9000 of 10002 . Current logp -4260.795 -4237.946 -4345.946 . Please wait!
Running DEzs-MCMC, chain 1 iteration 9300 of 10002 . Current logp -4151.762 -4199.456 -4282.755 . Please wait!
Running DEzs-MCMC, chain 1 iteration 9600 of 10002 . Current logp -4155.818 -4162.072 -4208.543 . Please wait!
Running DEzs-MCMC, chain 1 iteration 9900 of 10002 . Current logp -4155.818 -4107.946 -4195.48 . Please wait!
Running DEzs-MCMC, chain 1 iteration 10002 of 10002 . Current logp -4155.818 -4107.53 -4195.48 . Please wait!
Running DEzs-MCMC, chain 2 iteration 300 of 10002 . Current logp -629517.6 -923022.2 -2561376 . Please wait!
Running DEzs-MCMC, chain 2 iteration 600 of 10002 . Current logp -51551.69 -430489.4 -673674 . Please wait!
Running DEzs-MCMC, chain 2 iteration 900 of 10002 . Current logp -51551.69 -111994.2 -27651.78 . Please wait!
Running DEzs-MCMC, chain 2 iteration 1200 of 10002 . Current logp -14854.29 -90114.03 -22541.26 . Please wait!
Running DEzs-MCMC, chain 2 iteration 1500 of 10002 . Current logp -11954.81 -16117.86 -22541.26 . Please wait!
Running DEzs-MCMC, chain 2 iteration 1800 of 10002 . Current logp -10293.2 -9890.549 -15527.02 . Please wait!
Running DEzs-MCMC, chain 2 iteration 2100 of 10002 . Current logp -9592.388 -9748.39 -10280.27 . Please wait!
Running DEzs-MCMC, chain 2 iteration 2400 of 10002 . Current logp -9552.123 -9748.39 -9148.923 . Please wait!
Running DEzs-MCMC, chain 2 iteration 2700 of 10002 . Current logp -9142.205 -9249.991 -8784.532 . Please wait!
Running DEzs-MCMC, chain 2 iteration 3000 of 10002 . Current logp -8637.804 -9149.24 -8437.577 . Please wait!
Running DEzs-MCMC, chain 2 iteration 3300 of 10002 . Current logp -7403.693 -8433.703 -7564.009 . Please wait!
Running DEzs-MCMC, chain 2 iteration 3600 of 10002 . Current logp -7403.693 -7359.978 -7217.284 . Please wait!
Running DEzs-MCMC, chain 2 iteration 3900 of 10002 . Current logp -7394.26 -6964.411 -6923.753 . Please wait!
Running DEzs-MCMC, chain 2 iteration 4200 of 10002 . Current logp -7328.113 -6964.705 -6577.953 . Please wait!
Running DEzs-MCMC, chain 2 iteration 4500 of 10002 . Current logp -7300.692 -6371.044 -6436.208 . Please wait!
Running DEzs-MCMC, chain 2 iteration 4800 of 10002 . Current logp -7129.098 -5975.663 -5981.274 . Please wait!
Running DEzs-MCMC, chain 2 iteration 5100 of 10002 . Current logp -5994.924 -5883.562 -5303.358 . Please wait!
Running DEzs-MCMC, chain 2 iteration 5400 of 10002 . Current logp -5847.52 -5271.654 -4950.509 . Please wait!
Running DEzs-MCMC, chain 2 iteration 5700 of 10002 . Current logp -5322.339 -5137.526 -4950.509 . Please wait!
Running DEzs-MCMC, chain 2 iteration 6000 of 10002 . Current logp -5136.149 -5137.526 -4950.509 . Please wait!
Running DEzs-MCMC, chain 2 iteration 6300 of 10002 . Current logp -5093.861 -5137.526 -4820.956 . Please wait!
Running DEzs-MCMC, chain 2 iteration 6600 of 10002 . Current logp -5093.861 -4848.627 -4820.956 . Please wait!
Running DEzs-MCMC, chain 2 iteration 6900 of 10002 . Current logp -4820.472 -4773.136 -4820.956 . Please wait!
Running DEzs-MCMC, chain 2 iteration 7200 of 10002 . Current logp -4797.229 -4717.157 -4820.956 . Please wait!
Running DEzs-MCMC, chain 2 iteration 7500 of 10002 . Current logp -4797.229 -4717.157 -4743.401 . Please wait!
Running DEzs-MCMC, chain 2 iteration 7800 of 10002 . Current logp -4797.229 -4717.157 -4743.401 . Please wait!
Running DEzs-MCMC, chain 2 iteration 8100 of 10002 . Current logp -4754.817 -4717.157 -4727.38 . Please wait!
Running DEzs-MCMC, chain 2 iteration 8400 of 10002 . Current logp -4754.817 -4717.157 -4727.38 . Please wait!
Running DEzs-MCMC, chain 2 iteration 8700 of 10002 . Current logp -4753.639 -4717.157 -4717.157 . Please wait!
Running DEzs-MCMC, chain 2 iteration 9000 of 10002 . Current logp -4721.526 -4716.882 -4717.157 . Please wait!
Running DEzs-MCMC, chain 2 iteration 9300 of 10002 . Current logp -4707.452 -4716.882 -4715.054 . Please wait!
Running DEzs-MCMC, chain 2 iteration 9600 of 10002 . Current logp -4707.452 -4715.993 -4715.054 . Please wait!
Running DEzs-MCMC, chain 2 iteration 9900 of 10002 . Current logp -4701.201 -4715.288 -4715.054 . Please wait!
Running DEzs-MCMC, chain 2 iteration 10002 of 10002 . Current logp -4701.201 -4715.288 -4715.499 . Please wait! Check for convergence
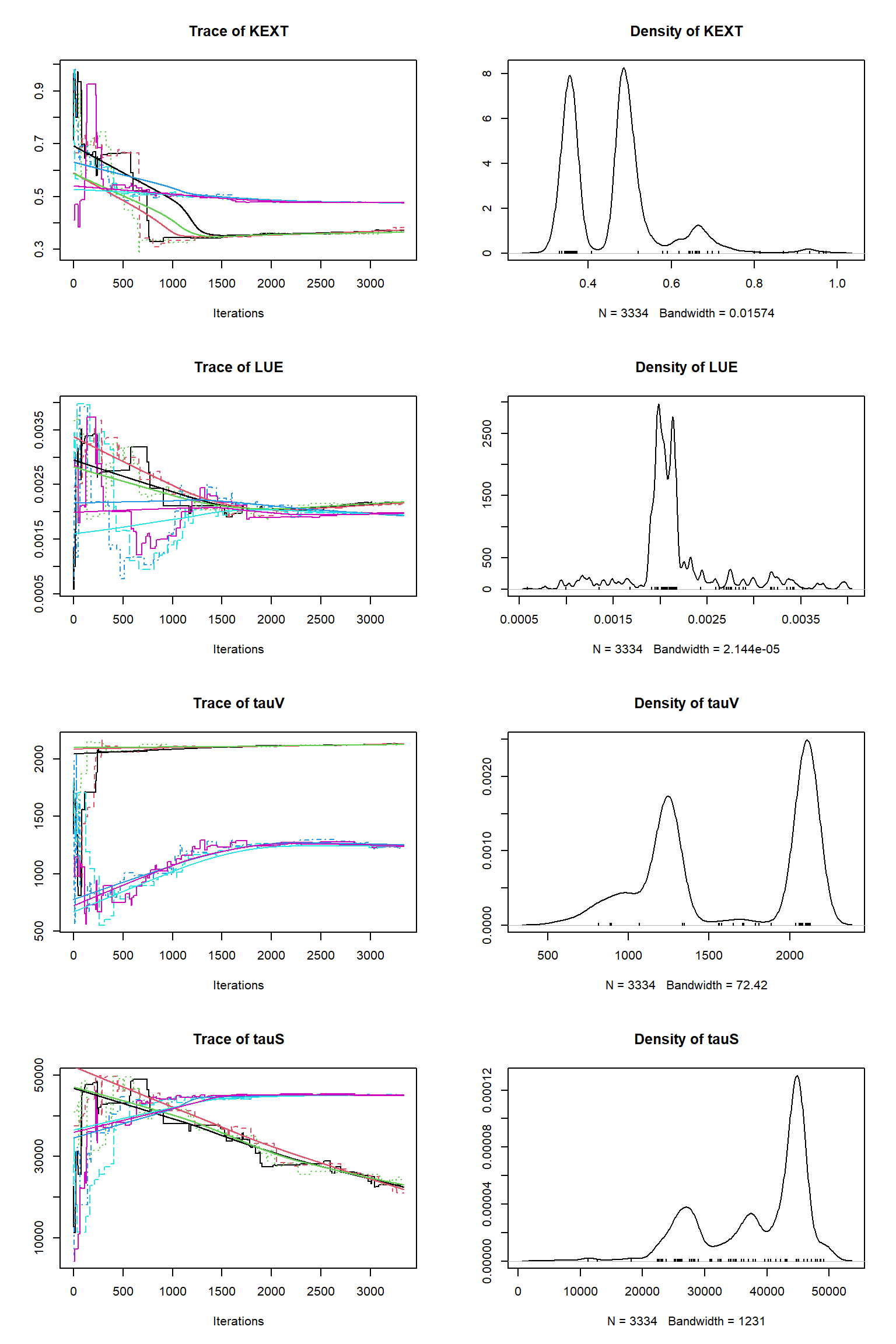
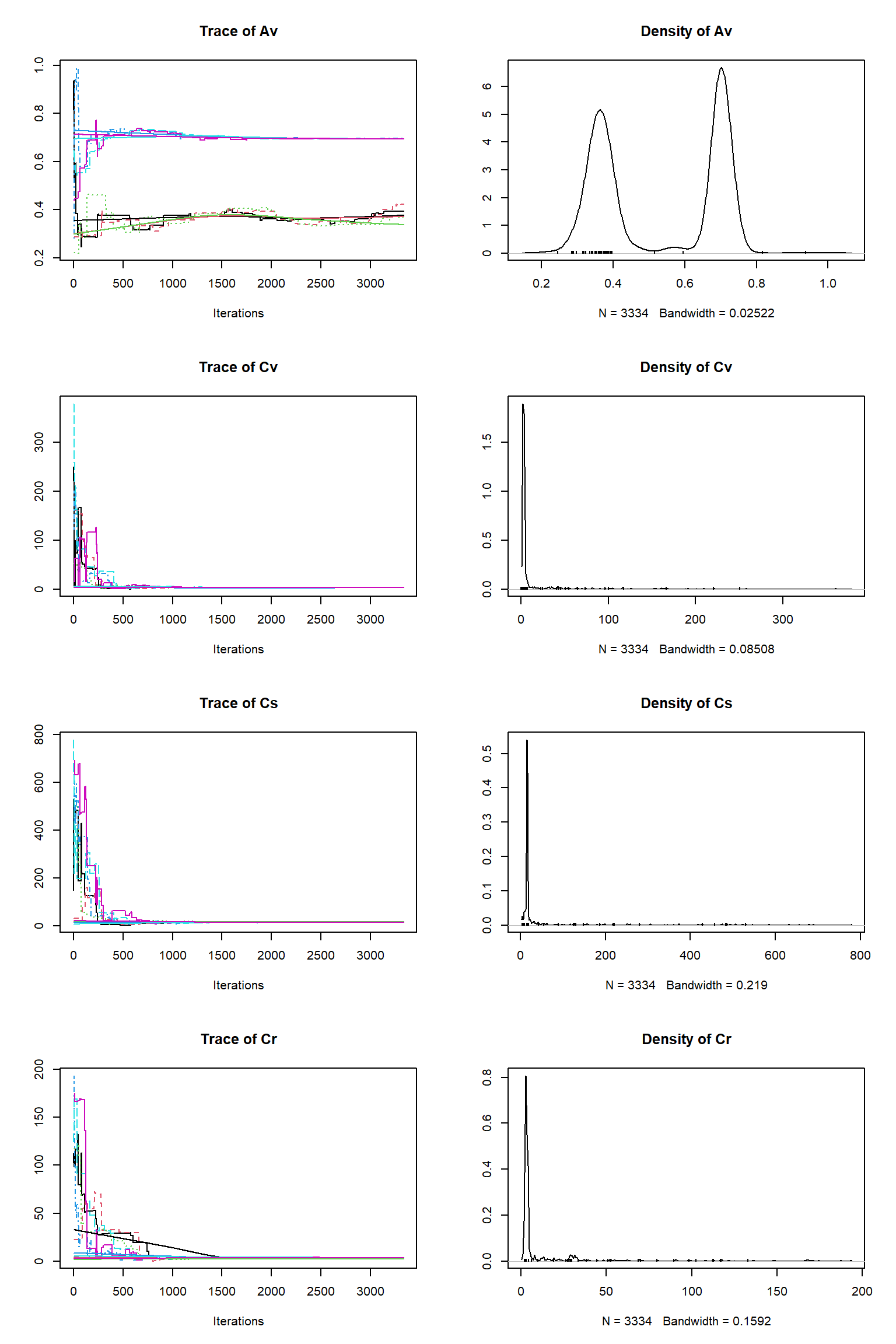
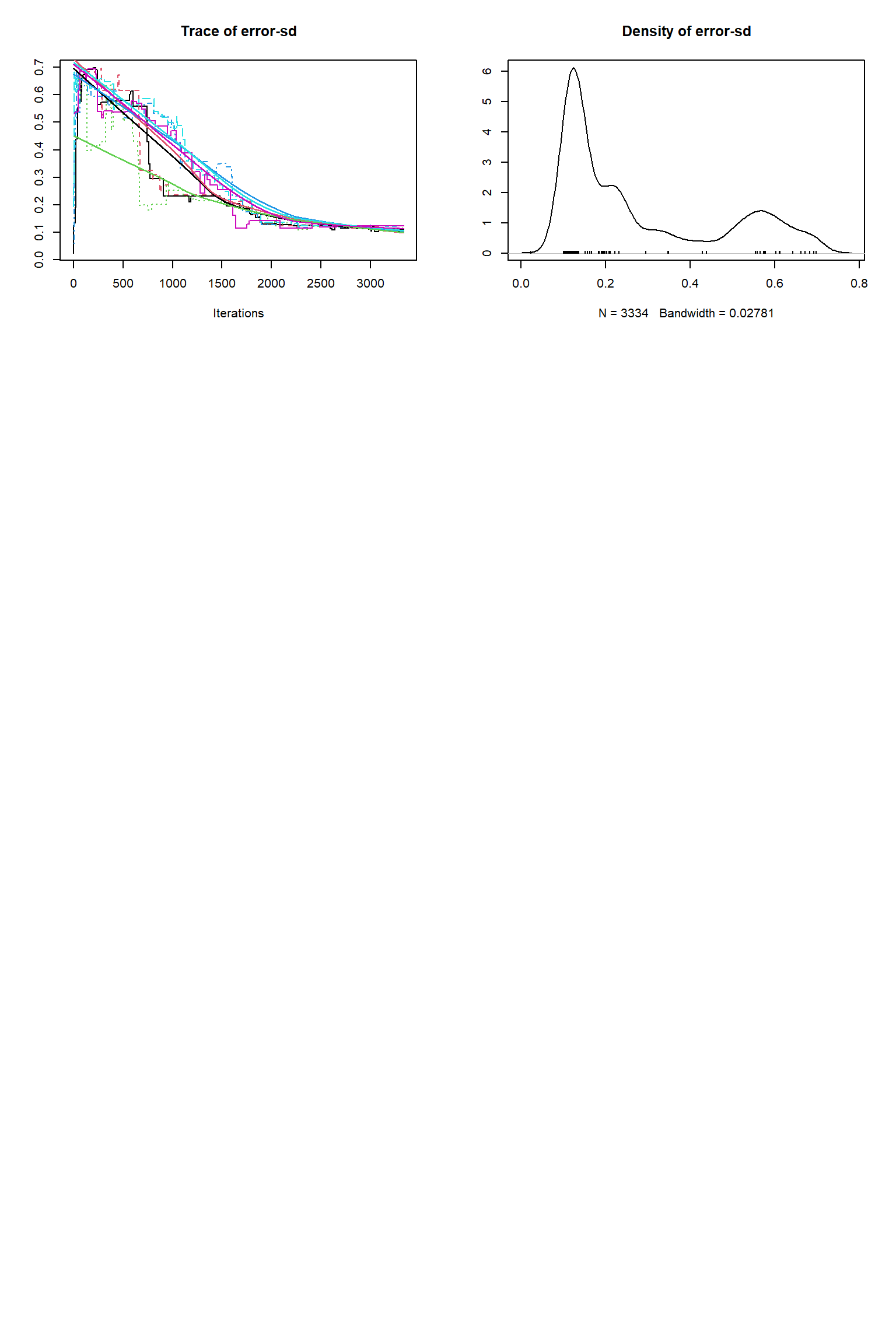
# # # # # # # # # # # # # # # # # # # # # # # # #
## MCMC chain summary ##
# # # # # # # # # # # # # # # # # # # # # # # # #
# MCMC sampler: DEzs
# Nr. Chains: 6
# Iterations per chain: 3334
# Rejection rate: 0.98
# Effective sample size: 90
# Runtime: 7.45 sec.
# Parameters
psf MAP 2.5% median 97.5%
KEXT 15.998 0.383 0.333 0.478 0.719
LUE 1.397 0.002 0.001 0.002 0.003
tauV 42.965 2132.579 782.149 1296.714 2129.069
tauS 4.917 21067.849 22538.699 42769.713 49108.707
Av 13.945 0.421 0.294 0.463 0.729
Cv 2.988 2.855 0.992 3.154 73.372
Cs 2.148 15.154 4.563 15.424 253.492
Cr 6.895 2.842 1.857 3.631 69.636
error-sd 1.035 0.105 0.109 0.198 0.684
## DIC: 1613306
## Convergence
Gelman Rubin multivariate psrf: 105.554
## Correlations
KEXT LUE tauV tauS Av Cv Cs Cr error-sd
KEXT 1.000 0.345 -0.564 0.292 0.393 0.522 0.440 0.527 0.684
LUE 0.345 1.000 0.228 -0.086 -0.409 0.357 0.366 0.477 0.353
tauV -0.564 0.228 1.000 -0.434 -0.903 -0.204 -0.211 -0.104 -0.352
tauS 0.292 -0.086 -0.434 1.000 0.486 -0.205 -0.331 -0.211 0.171
Av 0.393 -0.409 -0.903 0.486 1.000 -0.027 0.002 -0.120 0.081
Cv 0.522 0.357 -0.204 -0.205 -0.027 1.000 0.610 0.634 0.423
Cs 0.440 0.366 -0.211 -0.331 0.002 0.610 1.000 0.835 0.390
Cr 0.527 0.477 -0.104 -0.211 -0.120 0.634 0.835 1.000 0.525
error-sd 0.684 0.353 -0.352 0.171 0.081 0.423 0.390 0.525 1.000Check for convergence longer chain (150k)
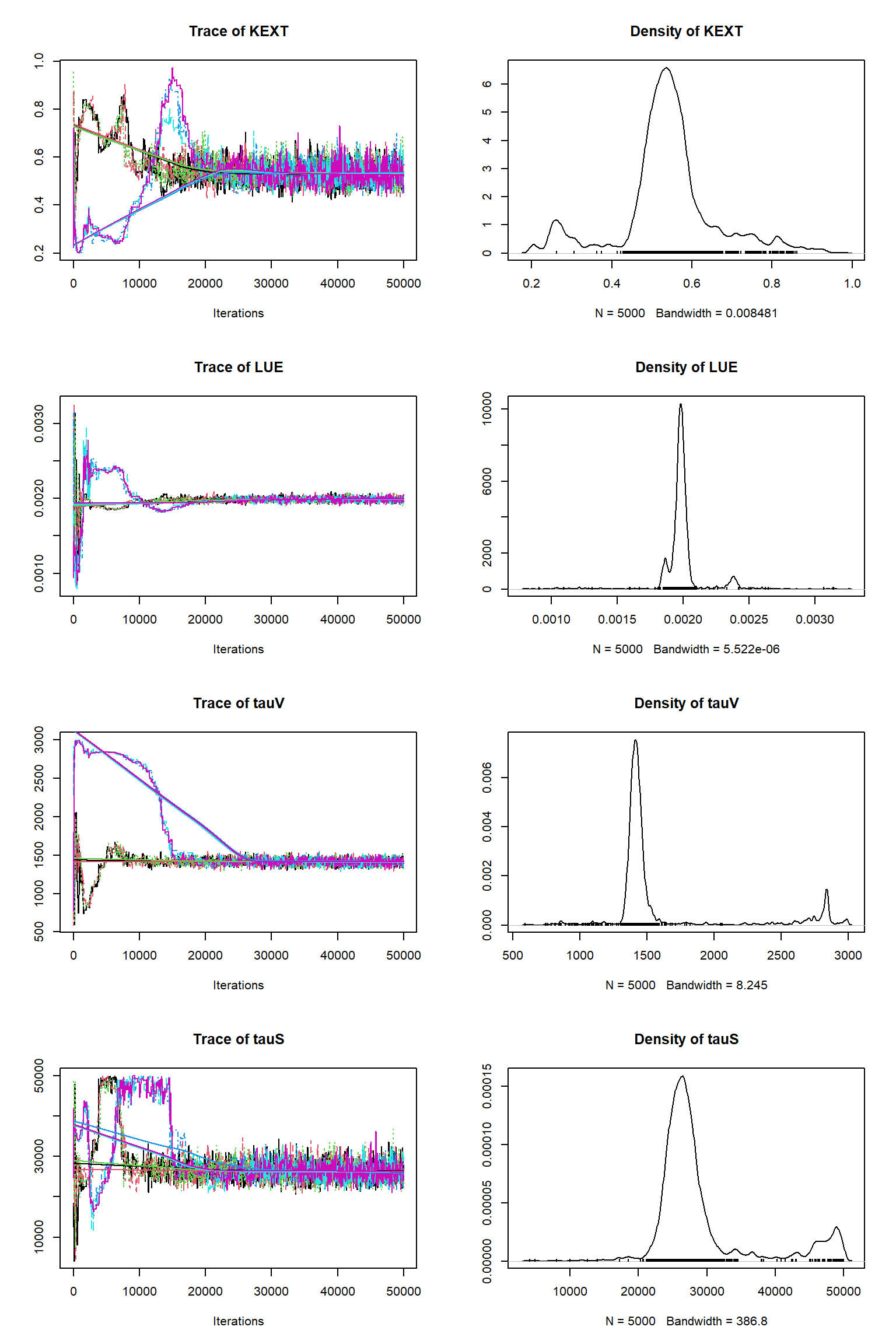
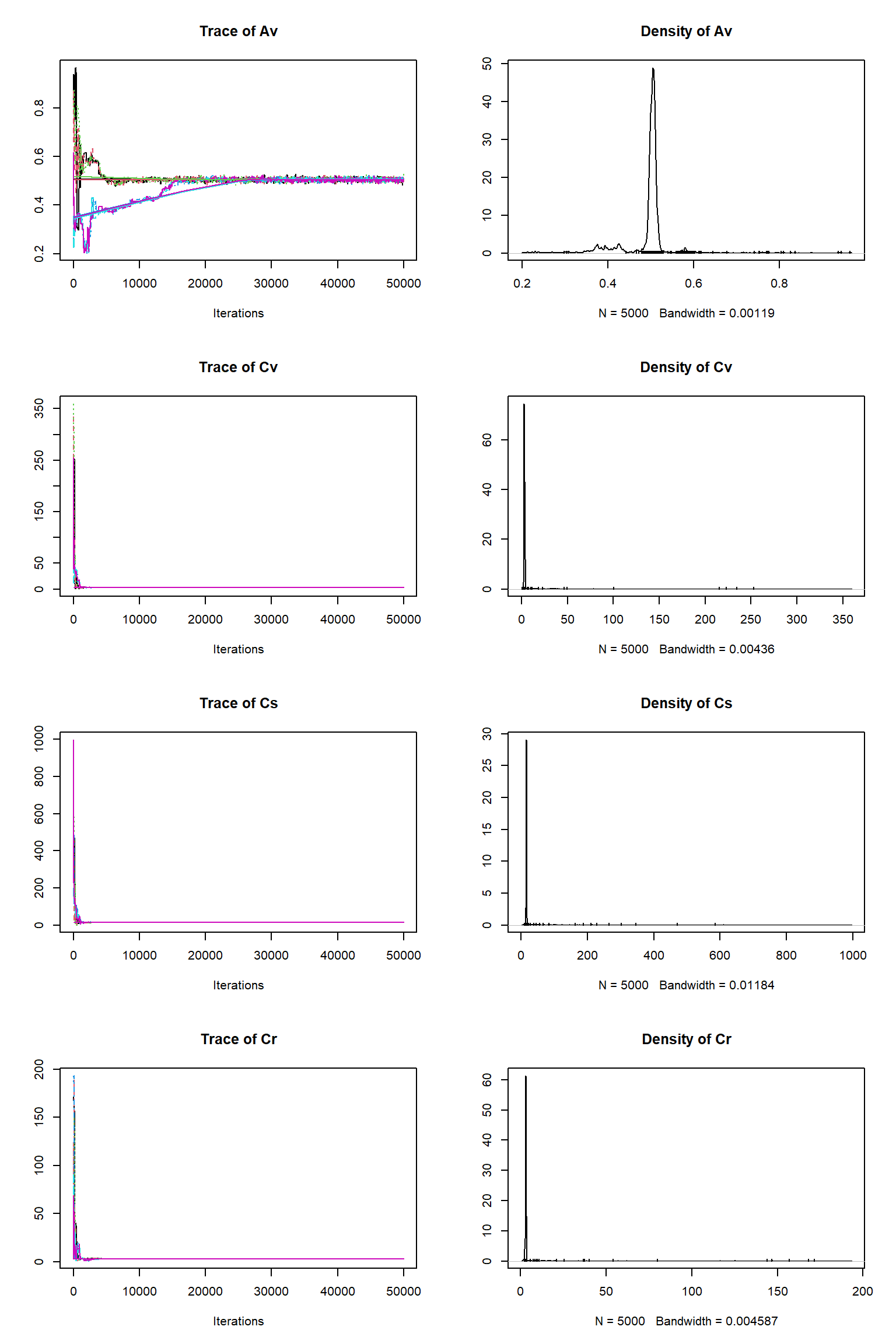

# # # # # # # # # # # # # # # # # # # # # # # # #
## MCMC chain summary ##
# # # # # # # # # # # # # # # # # # # # # # # # #
# MCMC sampler: DEzs
# Nr. Chains: 6
# Iterations per chain: 50000
# Rejection rate: 0.943
# Effective sample size: 719
# Runtime: 229.801 sec.
# Parameters
psf MAP 2.5% median 97.5%
KEXT 1.001 0.529 0.254 0.538 0.820
LUE 1.001 0.002 0.002 0.002 0.002
tauV 1.005 1394.867 1171.209 1425.719 2844.404
tauS 1.002 25672.714 21957.750 26787.986 49039.340
Av 1.008 0.508 0.355 0.503 0.578
Cv 1.002 3.020 2.849 3.008 3.460
Cs 1.007 14.915 14.503 14.952 16.571
Cr 1.009 3.037 2.648 3.025 3.371
error-sd 1.003 0.100 0.098 0.101 0.413
## DIC: 65745.51
## Convergence
Gelman Rubin multivariate psrf: 1.012
## Correlations
KEXT LUE tauV tauS Av Cv Cs Cr error-sd
KEXT 1.000 -0.386 -0.621 0.035 0.588 -0.035 -0.040 -0.019 -0.271
LUE -0.386 1.000 0.251 -0.112 -0.174 -0.079 -0.008 0.020 -0.248
tauV -0.621 0.251 1.000 0.507 -0.821 0.072 0.089 -0.002 0.367
tauS 0.035 -0.112 0.507 1.000 -0.371 0.014 0.045 -0.004 0.109
Av 0.588 -0.174 -0.821 -0.371 1.000 0.109 0.072 0.151 -0.263
Cv -0.035 -0.079 0.072 0.014 0.109 1.000 0.789 0.688 0.420
Cs -0.040 -0.008 0.089 0.045 0.072 0.789 1.000 0.711 0.484
Cr -0.019 0.020 -0.002 -0.004 0.151 0.688 0.711 1.000 0.444
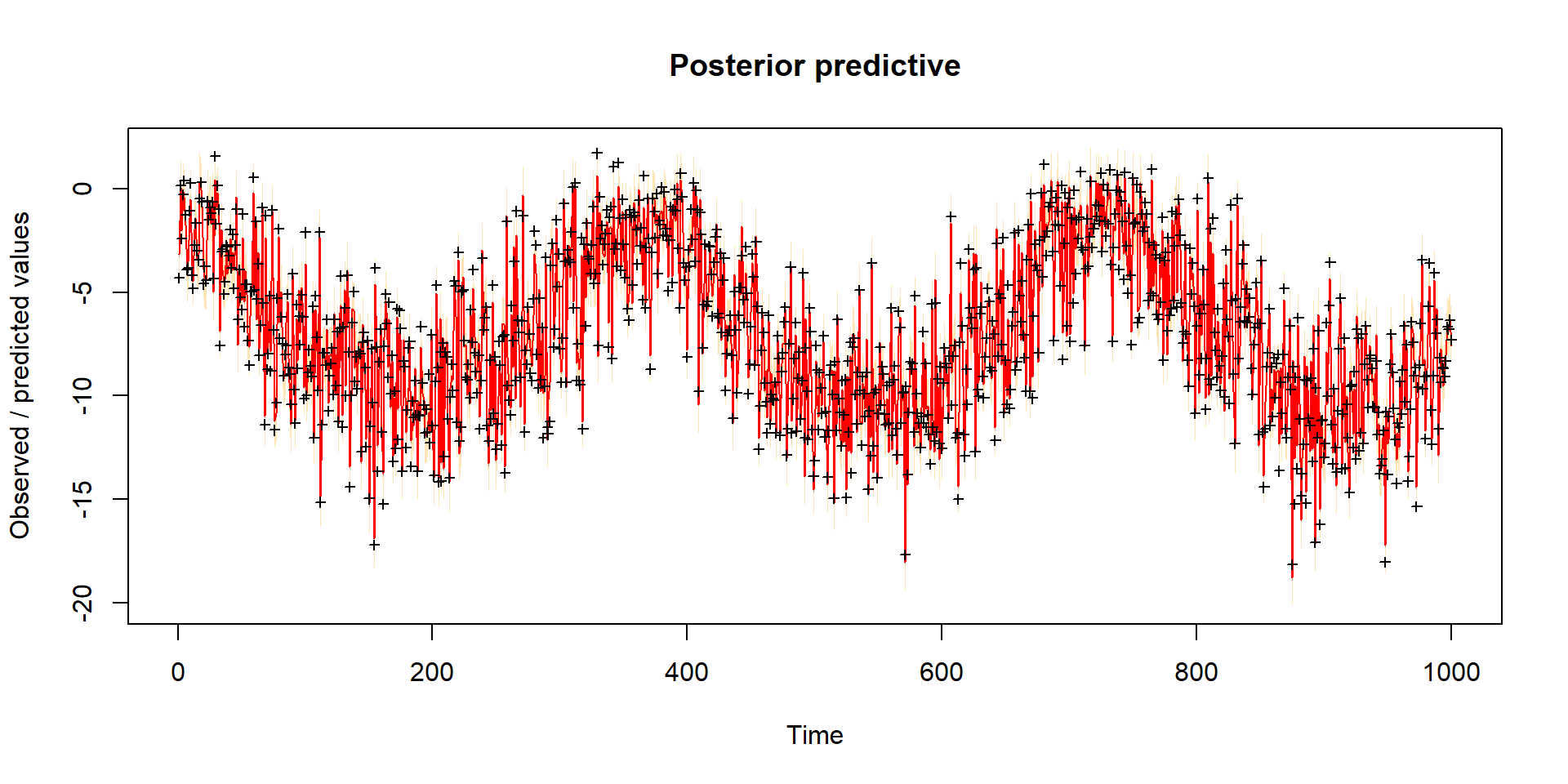
error-sd -0.271 -0.248 0.367 0.109 -0.263 0.420 0.484 0.444 1.000Remove burn-in and take sample
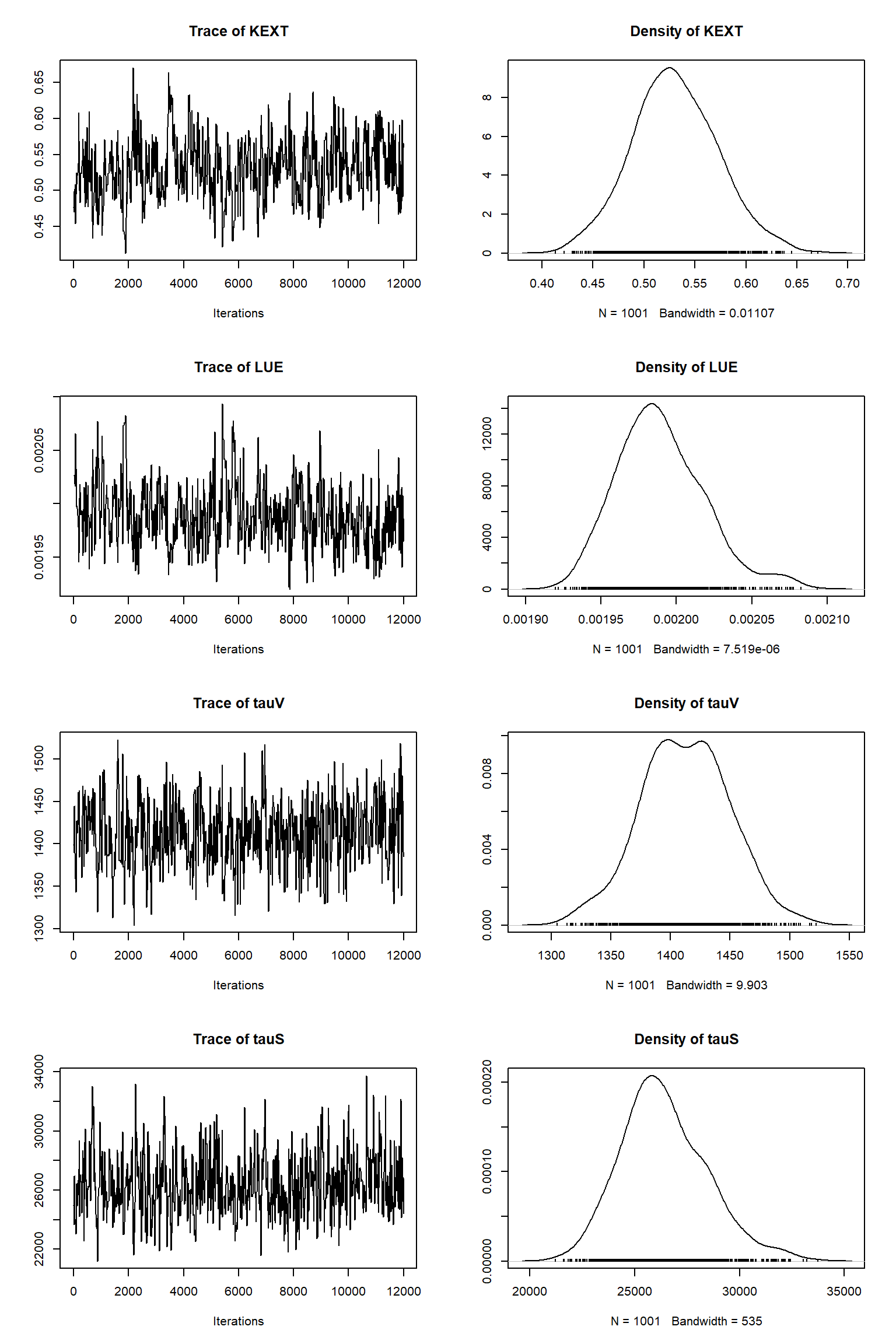
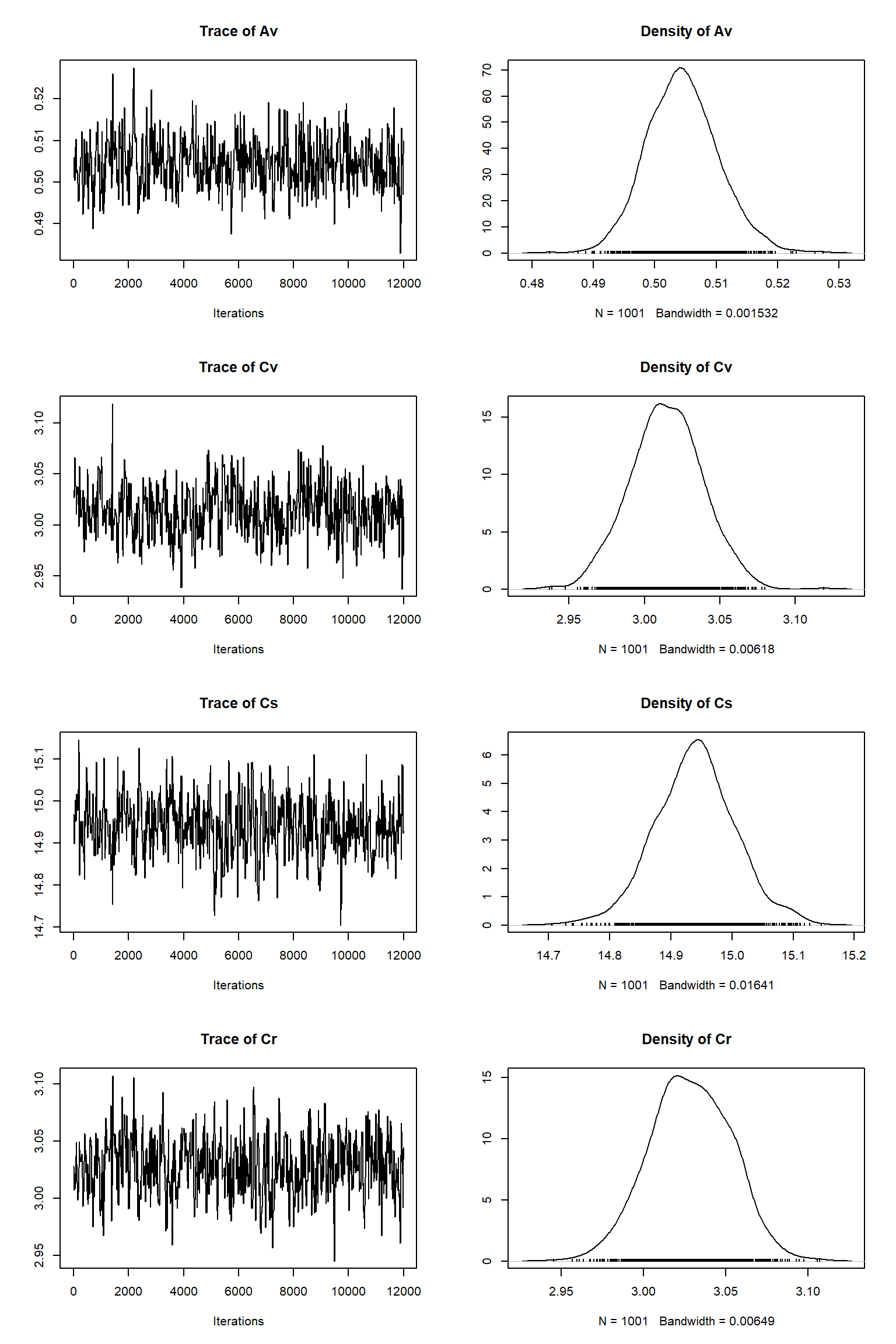

| best | lower | upper | |
|---|---|---|---|
| KEXT | 0.500 | 2e-01 | 1e+00 |
| LAR | 1.500 | 2e-01 | 3e+00 |
| LUE | 0.002 | 5e-04 | 4e-03 |
| GAMMA | 0.400 | 2e-01 | 6e-01 |
| tauV | 1440.000 | 5e+02 | 3e+03 |
| tauS | 27370.000 | 4e+03 | 5e+04 |
| tauR | 1440.000 | 5e+02 | 3e+03 |
| Av | 0.500 | 2e-01 | 1e+00 |
| Cv | 3.000 | 0e+00 | 4e+02 |
| Cs | 15.000 | 0e+00 | 1e+03 |
| Cr | 3.000 | 0e+00 | 2e+02 |
| error-sd | 0.100 | 1e-02 | 7e-01 |
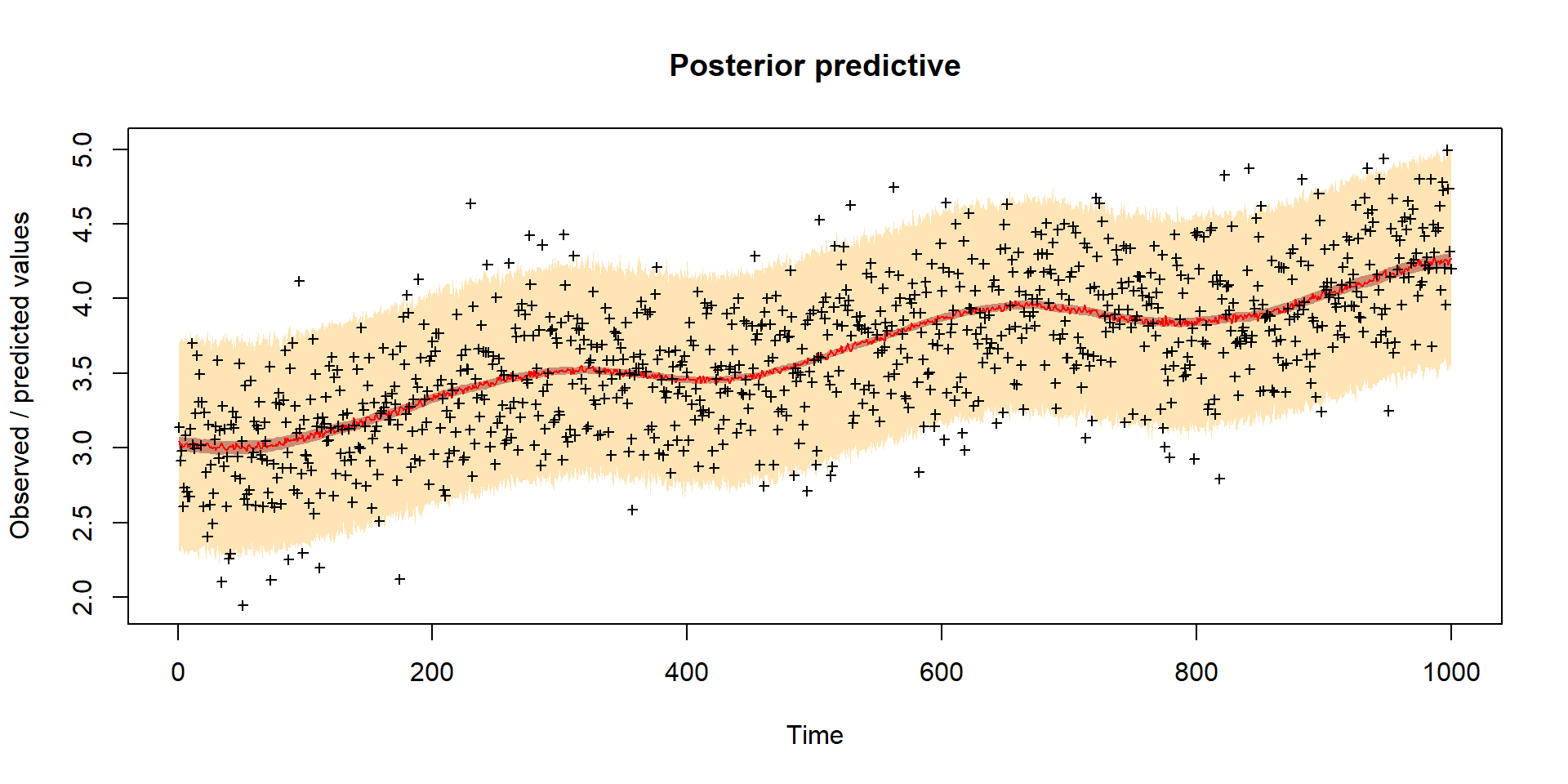
Model predictions
Computational Cost
VSEM is fast but even a “simple” model needed 150k runs
Much more complex “slower/expensive” models
- may need to be sped up
- can be replaced with statistical approximation (model emulation)
Sometime good enough to recode model in a more efficient language eg C or FORTRAN
More efficient inference schemes might be useful eg R package NIMBLE
Practical Session
We will now move onto a practical session
The web document for this can be accessed from the main page
Recap and Summary
What is a process based model PBM?
Why considering uncertainties in PBM predictions may be important
Sources of uncertainty in the Bayesian inference of PBMs
Worked example: Inference of a simple PBM (VSEM) using BayesianTools
Practical: Why it is important to represent all sources of uncertainty in Bayesian inference?