Several datasets are available to examine the ENM model performance and aid in model interpretation.
The performance measures can be retrieved from the
elements::PerformanceMeasures object; for example, below
the balanced accuracy from the random holdout sample and the model
tuning spatio-temporal cross-validation (Schratz et al., 2024) are
displayed.
pm_taxon <- subset(elements::PerformanceMeasures,
taxon_code == "stellaria_graminea",
select = c("Holdout.BalancedAccuracy", "STCV.BalancedAccuracy"))#> Holdout.BalancedAccuracy STCV.BalancedAccuracy
#> 8441 0.8579423 0.8001017The marginal effects of an ENM, in the form of Partial Dependency
Profile (PDP), Accumulated Local Effect (ALE) (Molnar, 2018), or
Hold-At-Optima (HOA) plots can also be viewed using the
elements::plot_me function. By setting the ‘presences’
argument to TRUE a box and whiskers plot showing the distribution of
presences is overlaid and by setting the ‘eivs’ argument to TRUE a point
and arrows showing the EIV and niche width values are overlaid, where
available in elements::VariableData.
elements::plot_me(taxa = "stellaria_graminea",
me_type = "ale",
free_y = TRUE,
presences = TRUE,
eivs = TRUE,
vars = c("L", "M", "N", "R", "S", "SD", "GP", "tmax_sm", "tmin_wt", "prec_wt", "prec_sm"))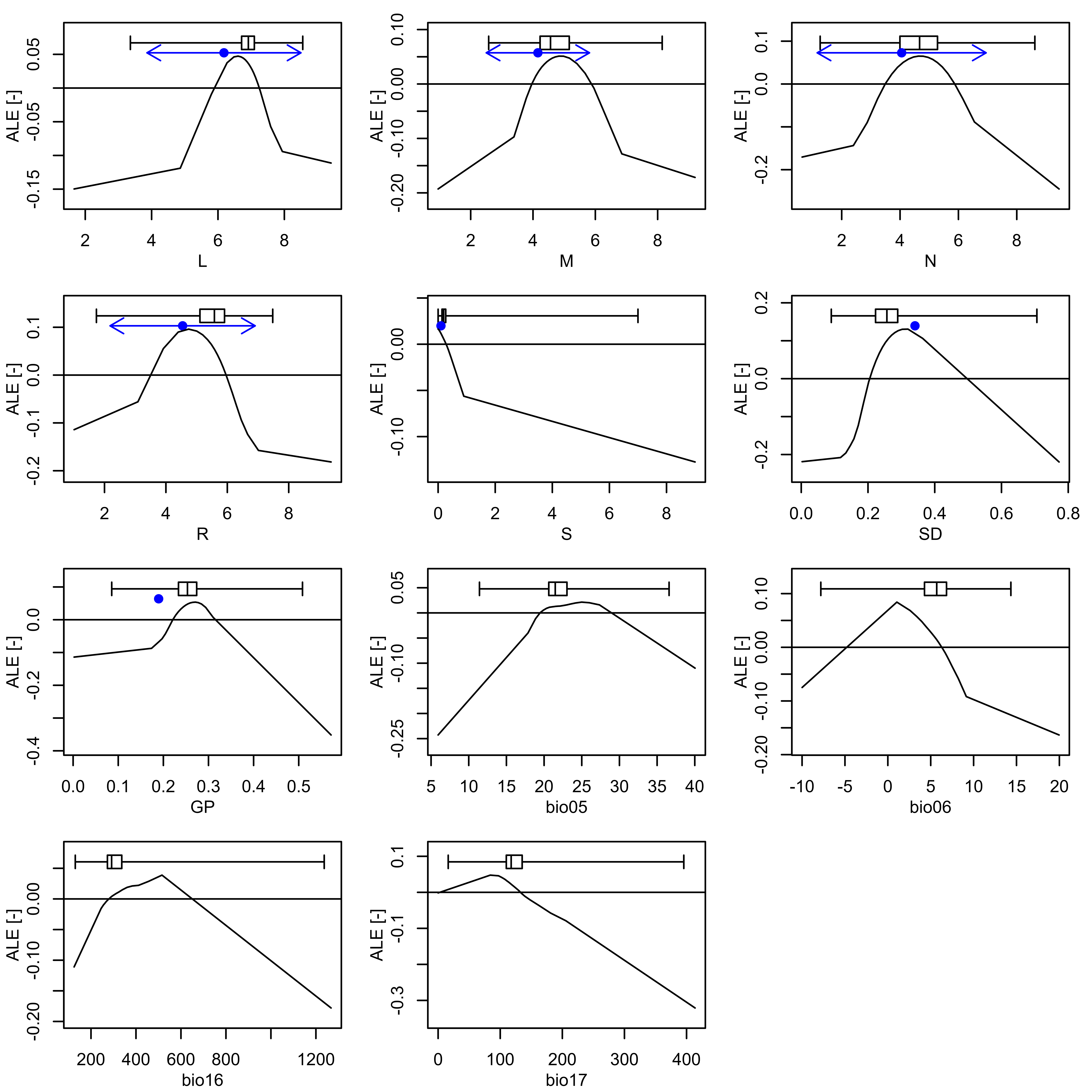
Multiple taxa can be supplied in the “taxa” argument, in this case the ability to plot presence box and whisker plots and EIV points and arrows is disabled.
elements::plot_me(taxa = c("galium_boreale", "galium_saxatile", "galium_uliginosum", "galium_sterneri", "galium_aparine"),
me_type = "pdp",
normalise = TRUE,
vars = c("L", "M", "N", "R", "S", "SD", "GP", "tmax_sm", "tmin_wt", "prec_wt", "prec_sm"))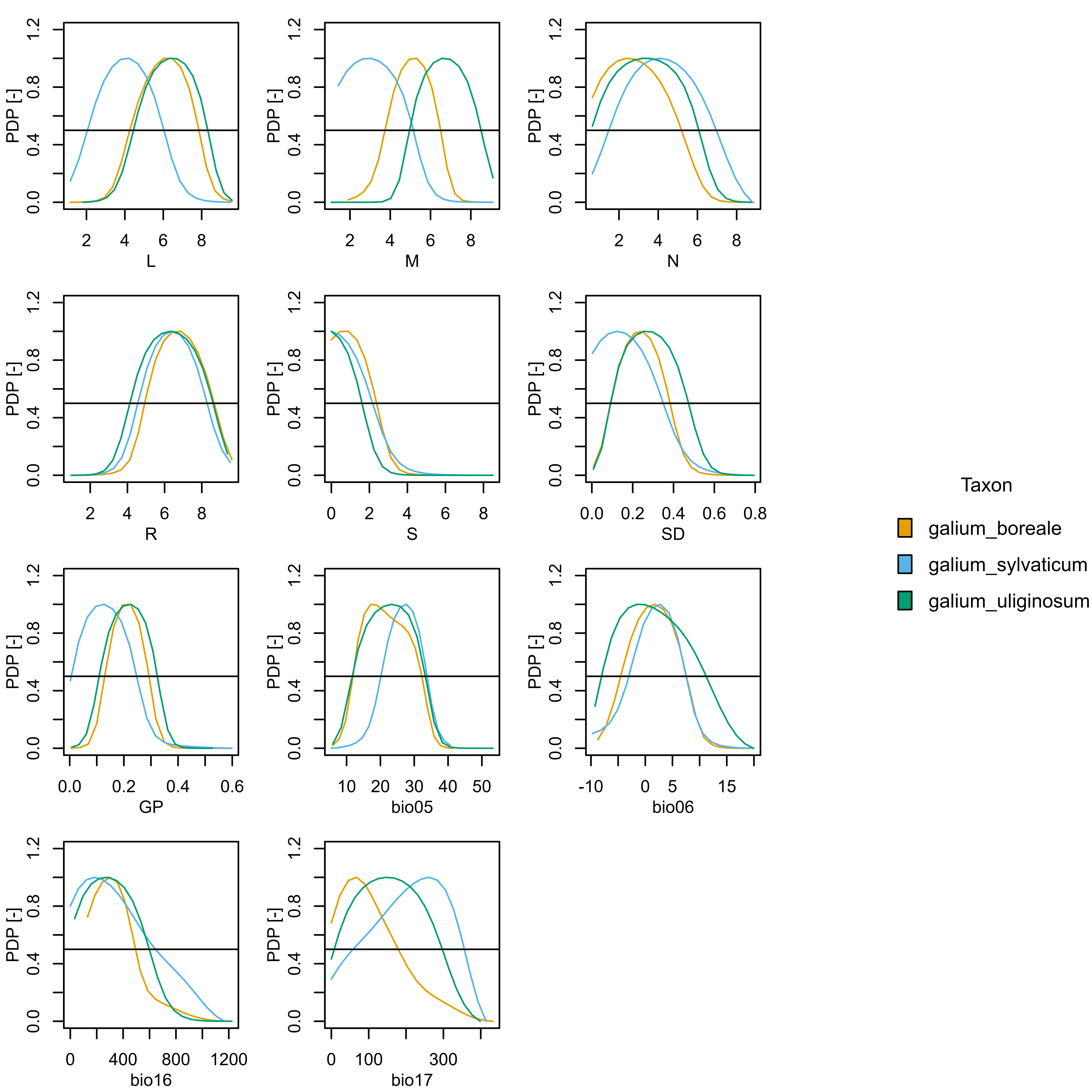
⚠ A note on ALE plots
In some instances the ALE curves may appear ‘inverted’ and not ecologically realistic, this is most often seen in situations where the distribution of presences along a variable gradient is extremely narrow and/or where there is a non-unimodal distribution, which causes extrapolation issues in the ALE calculations. For example, Torilis japonica has a extremely narrow distribution of plot-mean S values, with a maximum value of 3.25.
In these instances it is important to also visualise the PDP or HOA plots, which should then be prioritised when inspecting the shape of the univariate response.


</ details>