Retrieving Models
Due to the total size of the ENMs currently included in
elements (1.8GB when compressed, 7.5GB in memory) the ENMs
are not exported in a .rda object. Instead they are made available
through a filehash (Peng, 2005) database, which provides
access to the ENMs without loading all models into memory. To access the
ENMs a connection to this database must be initialised using
elements::startup. As mentioned above the Github repository
does not include the “./inst/extdata/Models” object containing all the
ENMs, the elements::startup will check whether the
“./inst/extdata/Models” is present and if it is not found will load the
“./inst/testdata/TestModels” models instead. The models to load can also
be accessed by passing “all” or “test” to the ‘models’ argument of
elements::startup.
elements::startup()
#> elements startup completed.
model <- elementsEnv$Models[["stellaria_graminea"]]#>
#> Call:
#> svm(formula = Presence ~ L + M + N + R + S + SD + GP + tmax_sm +
#> tmin_wt + prec_sm + prec_wt, data = data, type = "C-classification",
#> probability = TRUE)
#>
#>
#> Parameters:
#> SVM-Type: C-classification
#> SVM-Kernel: radial
#> cost: 0.23
#>
#> Number of Support Vectors: 30545Generating Predictions
The raw ENMs retrieved using the method above can be used as regular
e1071 SVM model objects. Alternatively, the helper function
elements::predict_occ_taxon retrieves a model using the
method above, generates predictions, and formats the results as a data
frame.
results <- elements::predict_occ_taxon(taxon = "stellaria_graminea", predictors = elements::ExampleData1, pa = "Present", limit = NULL, dp = 2, append = "ids")#> Present
#> 1 0.03
#> 2 0.25
#> 3 0.06
#> 4 0.08
#> 5 0.07
#> 6 0.74An additional helper function elements::predict_occ can
generate predictions for multiple taxa, by either specifing the taxa to
model in the ‘taxa’ argument, or by setting ‘taxa’ to NULL and including
an additional column in the predictors data frame named
‘taxon_code’.
results <- elements::predict_occ(taxa = NULL, predictors = elements::ExampleData2, pa = "Present", limit = NULL, holdopt = NULL, dp = 2, append = "ids")#> taxon_code Present
#> 201 silene_flos-cuculi 0.01
#> 202 silene_flos-cuculi 0.02
#> 203 silene_flos-cuculi 0.04
#> 204 silene_flos-cuculi 0.02
#> 205 silene_flos-cuculi 0.01
#> 206 silene_flos-cuculi 0.08Two helper arguments provide additional functionality in controlling
model use. First, is the ‘limit’ argument, which assigns probability
values of zero if one or more predictors are outside a specified range
e.g. the 10% and 90% quantiles (see elements::NicheWidths).
Second, is the ‘holdopt’ argument, which holds specified variable values
at their optima (as defined by the mean value present in
elements::NicheWidths).
As a simple demonstration, below two sets of predictions for Stellaria graminea are generated, holding all variables apart from N at their optima: 1) with no limit set, and 2) with a limit set to the 1% and 99% quantiles.
n_gradient <- data.frame("N" = seq(0, 10, 0.01))
vary_N_no_limit <- elements::predict_occ_taxon(taxon = "stellaria_graminea", predictors = n_gradient,
pa = "Present", limit = NULL, holdopt = c("tmax_sm", "tmin_wt", "prec_wt", "prec_sm", "GP", "L", "M", "R", "S", "SD"),
dp = 2, append = "predictors")
vary_N_q01_q99 <- elements::predict_occ_taxon(taxon = "stellaria_graminea", predictors = n_gradient,
pa = "Present", limit = "q01_q99", holdopt = c("tmax_sm", "tmin_wt", "prec_wt", "prec_sm", "GP", "L", "M", "R", "S", "SD"),
dp = 2, append = "predictors")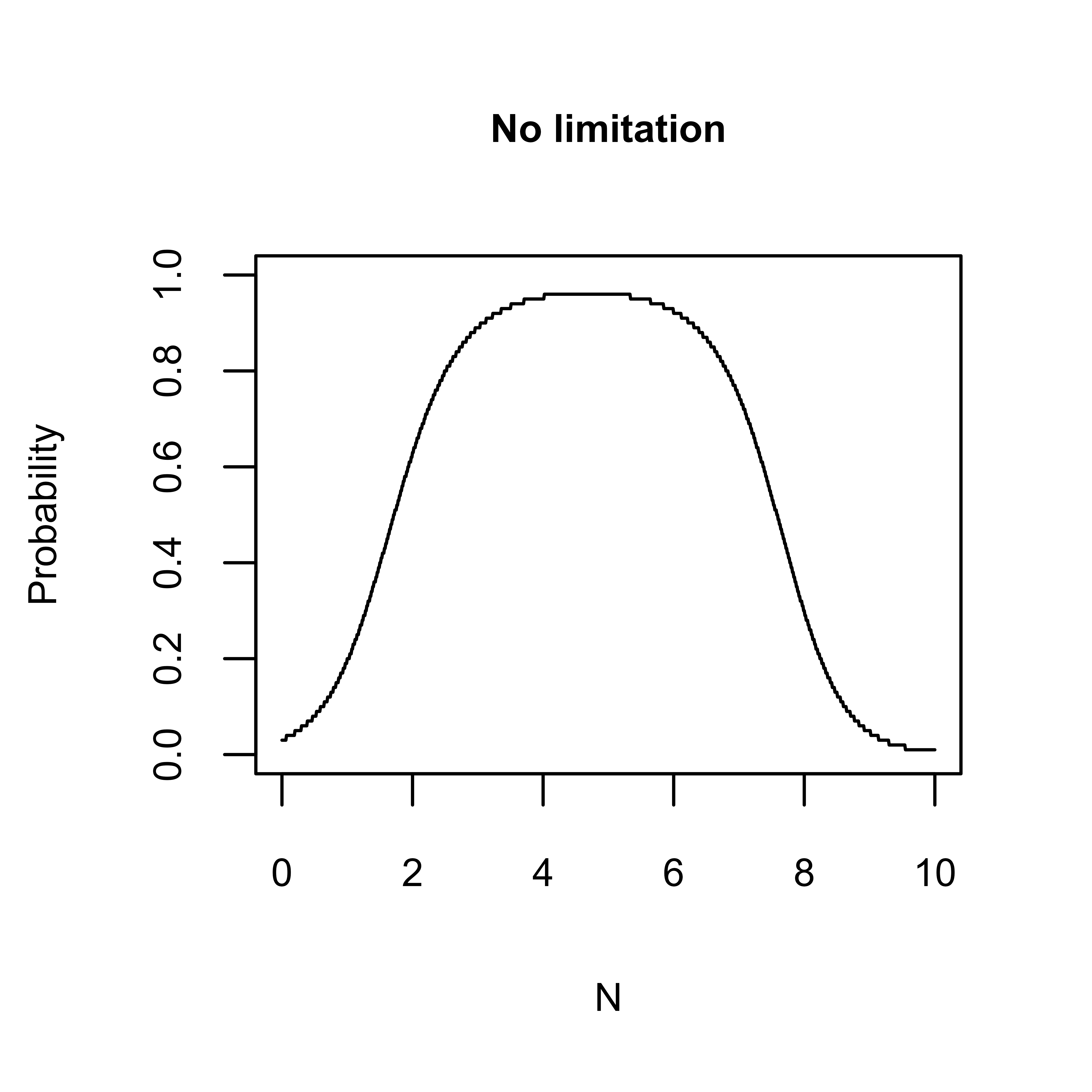
Please note that as ten out of the eleven variables are held at their
optima the predicted probabilities will be high as the influence of
unsuitable N values will be partially offset. Consequently, the shape of
the response curves above will be wider than the corresponding PDP plot
produced with the elements::plot_me function (see the
Model Inspection).
Environmental filtering
elements can also be used to filter species pools based
on a given set of predictor values using the function
elements::env_filter. Two sets of methods are available: 1)
“svm” which generates predictions using
elements::predict_occ and uses the resultant probability
values; and 2) “mean” and “median” which calculates the normalised
euclidean distance between the values supplied in the ‘predictors’
argument and the mean or median niche positions as present in
elements::NicheWidths.
NOTE: The mean and median methods are only included for demonstrative purposes only and should not be used in practice as they do not consider the joint distribution of variables as expressed through the SVM model hypervolumes.
The option to apply the elements::envelope_filter
function, which first screens the supplied taxa to check whether the
predictor values are within a given range as supplied to the ‘limit’
argument (“min_max”, “q01_q99”, “q05_q95”, “q10_q90”, “q25_q75”), is
controlled by the ‘screen’ argument; elements::env_filter
will then only run the more computationally expensive
elements::predict_occ or
elements::calc_distance functions for the taxa within the
limits. This is highly recommended as it usually results in a 95%
reduction in the number of taxa being supplied to
elements::predict_occ or
elements::calc_distance, greatly improving performance.
For example, below elements::env_filter is applied to
all taxa in elements::ModelledTaxaCodes using the svm
method, with the predictors derived from
elements::ExampleScenarios.
filter_results <- elements::env_filter(predictors = elements::ExampleScenarios[1,],
taxa = elements::ModelledTaxaCodes,
screen = TRUE, method = "svm", limit = "min_max",
exclude = NULL, threshold = NULL,
append = "ids")#> scenario_code scenario_stage climate_scenario timeslice scenario_name
#> 1 a.i baseline historical 2007 Climate Change
#> 2 a.i baseline historical 2007 Climate Change
#> 3 a.i baseline historical 2007 Climate Change
#> 4 a.i baseline historical 2007 Climate Change
#> 5 a.i baseline historical 2007 Climate Change
#> 6 a.i baseline historical 2007 Climate Change
#> 7 a.i baseline historical 2007 Climate Change
#> 8 a.i baseline historical 2007 Climate Change
#> 9 a.i baseline historical 2007 Climate Change
#> 10 a.i baseline historical 2007 Climate Change
#> taxon_code Present
#> 1 abies_alba 0.021
#> 2 abietinella_abietina 0.020
#> 3 acer_campestre 0.037
#> 4 acer_negundo 0.005
#> 5 acer_platanoides 0.033
#> 6 acer_pseudoplatanus 0.046
#> 7 acer_tataricum 0.002
#> 8 achillea_millefolium 0.068
#> 9 achillea_ptarmica 0.322
#> 10 aconogonon_alpinum 0.002In practice, given the limited size of regional species pools and/or
interest in a particular group of taxa only it is often more practical
to supply a reduced list of taxa to
elements::env_filter.
Niche overlap
The ability to calculate the niche overlap between taxa is provided
through the elements::calc_overlap function, below the
niche overlaps for the taxa present in
elements::ExamplePlot are displayed.
elements::calc_overlap(taxa = elements::ExamplePlot$taxon_code)
#> taxon_code_1 taxon_code_2 GP
#> 1 carex_rostrata equisetum_fluviatile 0.7214768
#> 2 carex_rostrata galium_palustre_subsp._palustre 0.8363951
#> 3 carex_rostrata hydrocotyle_vulgaris 0.8881867
#> 4 carex_rostrata juncus_effusus 0.7757002
#> 5 carex_rostrata silene_flos-cuculi 0.7454230
#> 6 equisetum_fluviatile galium_palustre_subsp._palustre 0.7898207
#> 7 equisetum_fluviatile hydrocotyle_vulgaris 0.7940598
#> 8 equisetum_fluviatile juncus_effusus 0.9170747
#> 9 equisetum_fluviatile silene_flos-cuculi 0.5435305
#> 10 galium_palustre_subsp._palustre hydrocotyle_vulgaris 0.8869708
#> 11 galium_palustre_subsp._palustre juncus_effusus 0.8073129
#> 12 galium_palustre_subsp._palustre silene_flos-cuculi 0.6764641
#> 13 hydrocotyle_vulgaris juncus_effusus 0.7661815
#> 14 hydrocotyle_vulgaris silene_flos-cuculi 0.6705272
#> 15 juncus_effusus silene_flos-cuculi 0.6206947
#> L M N prec_sm prec_wt R S
#> 1 0.7019289 0.7653469 0.7580541 0.7176748 0.6973665 0.8166178 0.7107549
#> 2 0.7453617 0.8966472 0.8877305 0.6650737 0.6928014 0.9209143 0.8004903
#> 3 0.8583172 0.8997119 0.8192811 0.8803404 0.6878805 0.7909851 0.6780486
#> 4 0.6630992 0.7802450 0.7328599 0.7242175 0.8028249 0.8776274 0.9099244
#> 5 0.8016116 0.6011772 0.7273055 0.8601537 0.8501649 0.7312728 0.8835696
#> 6 0.8603687 0.7431525 0.7500407 0.5177036 0.4843814 0.7957846 0.6828651
#> 7 0.8159822 0.7956703 0.6578178 0.7196383 0.9497895 0.9453407 0.5714592
#> 8 0.8518272 0.7689862 0.8182046 0.9794560 0.8724973 0.7291969 0.7011042
#> 9 0.5847399 0.4424183 0.5340497 0.8231144 0.7904267 0.6222805 0.7425152
#> 10 0.8275578 0.9348687 0.7156526 0.7474427 0.4821835 0.7561528 0.8451622
#> 11 0.8116754 0.8401339 0.8229316 0.5252077 0.5704317 0.8127146 0.7111169
#> 12 0.6468140 0.6382254 0.7093616 0.6213255 0.6412937 0.7721620 0.7261270
#> 13 0.7074216 0.8769743 0.5664275 0.7183427 0.8718478 0.7442617 0.5947429
#> 14 0.7220341 0.6106629 0.6729223 0.8382208 0.7762921 0.5809344 0.6147410
#> 15 0.5672006 0.5883891 0.6175172 0.8221031 0.8292024 0.6390688 0.9058863
#> SD tmax_sm tmin_wt
#> 1 0.6158546 0.5878702 0.6209539
#> 2 0.8161466 0.5504271 0.6054813
#> 3 0.6543901 0.7730851 0.7574574
#> 4 0.7287424 0.8100591 0.8836285
#> 5 0.6906799 0.8616340 0.8790914
#> 6 0.6765197 0.3324481 0.3615850
#> 7 0.8617632 0.6452016 0.5806407
#> 8 0.8330254 0.7166949 0.5909182
#> 9 0.4890992 0.6427807 0.6376136
#> 10 0.7678227 0.4476872 0.5183362
#> 11 0.8011935 0.4468069 0.6051843
#> 12 0.6733674 0.4586788 0.5356794
#> 13 0.9241939 0.7829824 0.8560658
#> 14 0.5518042 0.8428999 0.7555202
#> 15 0.6126248 0.8808119 0.8054637Shutting down
At the end of the analysis run elements::shutdown to
close the connection to the filehash database.
elements::shutdown()